Visualise hit guides
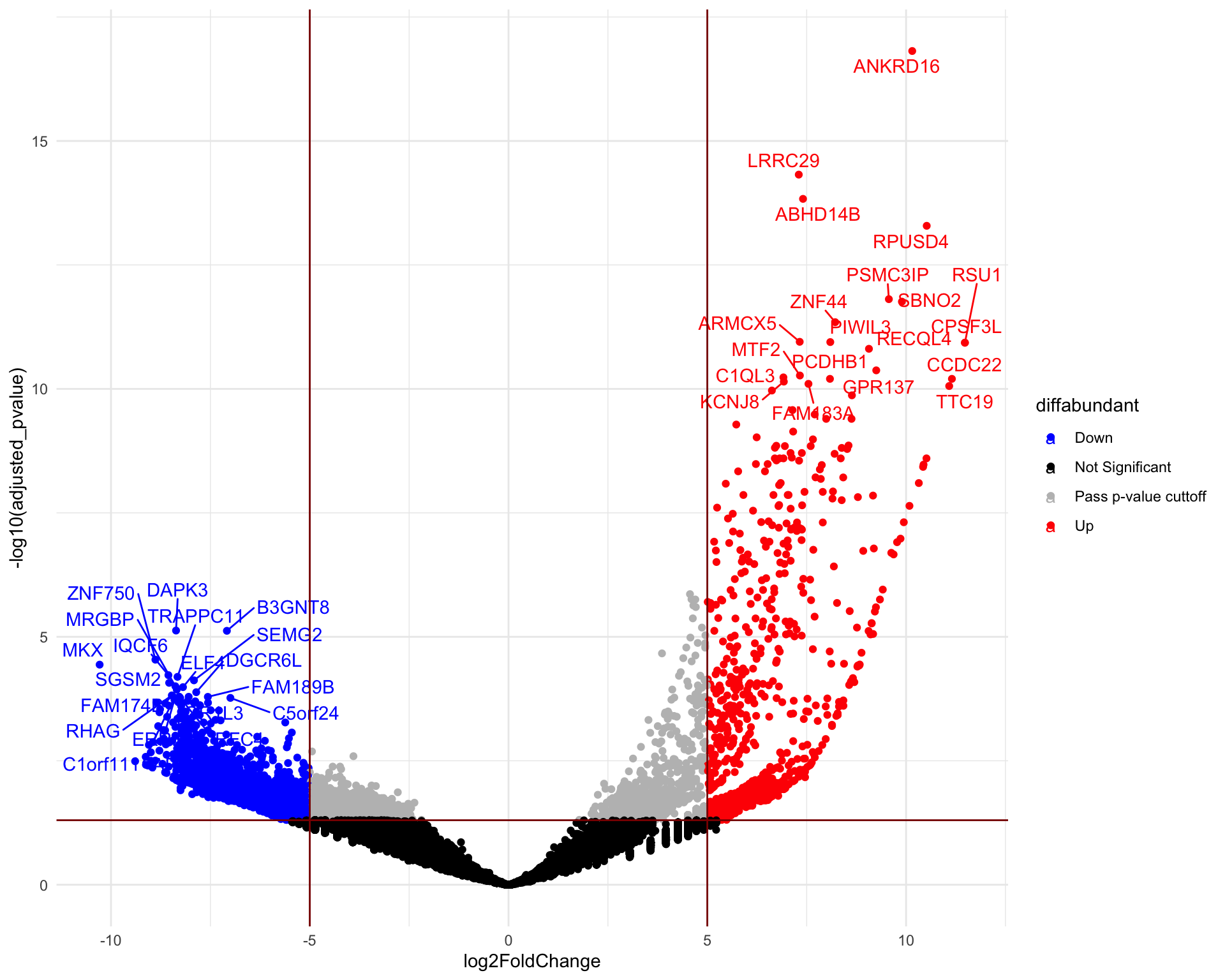
A volcano plot is a scatter plot that visualizes the differential abundance of guides. The fold change is typically displayed on the x-axis, while the y-axis represents the corresponding p-values. The significantly differentially abundant genes are the ones found in the upper-left and upper-right corners. Below we visualise the hits for ToxA vs Control and ToxB vs Control samples, respectively.
Volcano plot (ToxA vs Control)
# remove rows that contain NA values
de <- lrt2.toxa[complete.cases(lrt2.toxa), ]
colnames(de)[colnames(de) == "logFC"] <- "log2FoldChange"
colnames(de)[colnames(de) == "PValue"] <- "pvalue"
colnames(de)[colnames(de) == "Symbol"] <- "gene_symbol"
colnames(de)[colnames(de) == "FDR"] <- "adjusted_pvalue"
de$diffabundant <- "Pass p-value cuttoff"
de$diffabundant[de$adjusted_pvalue > 0.05] <- "Not Significant"
de$diffabundant[de$log2FoldChange < -5 & de$log2FoldChange >= 5 & de$adjusted_pvalue <
0.05] <- "Pass p-value & Log2FC cuttoff"
de$diffabundant[de$log2FoldChange >= 5 & de$adjusted_pvalue <= 0.05] <- "Up"
de$diffabundant[de$log2FoldChange < -5 & de$adjusted_pvalue <= 0.05] <- "Down"
head(de) gene_symbol UID seq log2FoldChange
HGLibB_38324 PRDM13 HGLibB_38324 GCAAGTACCTGTCAGACCGC 11.209176
HGLibA_32966 NUDT2 HGLibA_32966 ATGAGCACCAAGCCTACCGC 11.594388
HGLibB_42762 SBSPON HGLibB_42762 GACAAGCTACGTCTCCACAC 12.255728
HGLibB_44908 SLC35B2 HGLibB_44908 TCCGCCTGAAGTACTGCACC 10.286574
HGLibB_45454 SLIT2 HGLibB_45454 TCTCTAGTTCTTTAAGATCC 11.404868
HGLibB_23505 IQGAP3 HGLibB_23505 CACTCACAGGCTGCCCAGCC 9.781491
logCPM LR pvalue adjusted_pvalue diffabundant
HGLibB_38324 10.829302 89.05910 3.831913e-21 2.143074e-16 Up
HGLibA_32966 8.780164 73.01061 1.289546e-17 3.195504e-13 Up
HGLibB_42762 9.530267 72.44893 1.714112e-17 3.195504e-13 Up
HGLibB_44908 11.311403 69.70313 6.893661e-17 9.638544e-13 Up
HGLibB_45454 8.763229 68.20288 1.475103e-16 1.649962e-12 Up
HGLibB_23505 9.434533 66.97834 2.745064e-16 2.491273e-12 Upg_down <- which(de$log2FoldChange > 5 & de$adjusted_pvalue <= 0.05)
g_up <- which(de$log2FoldChange < -5 & de$adjusted_pvalue <= 0.05)
de$delabel <- NA
de$delabel[c(g_down[1:20], g_up[1:20])] <- de$gene_symbol[c(g_down[1:20], g_up[1:20])]
ggplot(data = de, aes(x = log2FoldChange, y = -log10(adjusted_pvalue), col = diffabundant,
label = delabel)) + geom_point() + theme_minimal() + geom_text_repel(max.overlaps = Inf) +
scale_color_manual(values = c("blue", "black", "gray", "red")) + geom_vline(xintercept = c(-5,
5), col = "darkred") + geom_hline(yintercept = -log10(0.05), col = "darkred")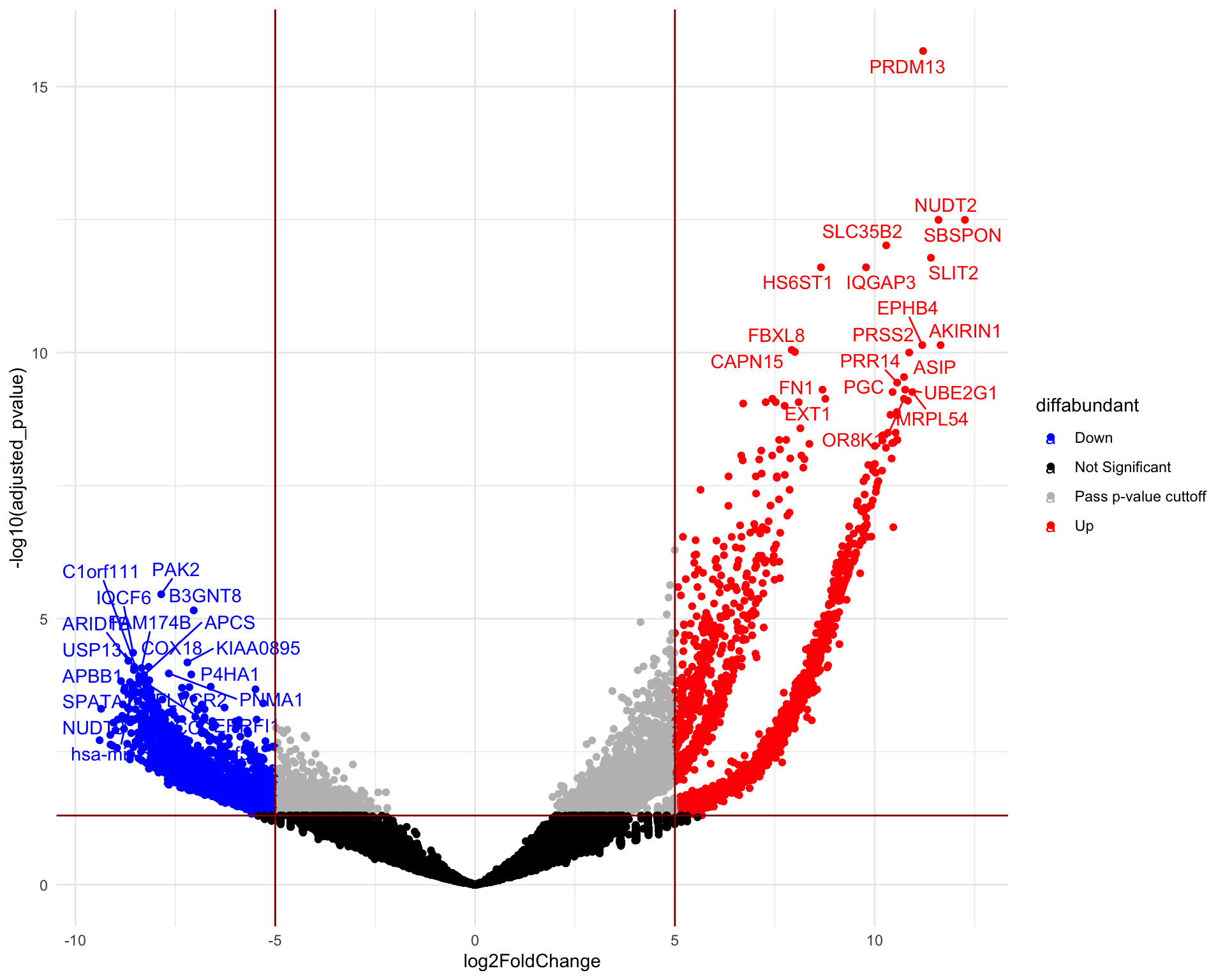
Volcano plot (ToxB vs Control)
# remove rows that contain NA values
de <- lrt2.toxb[complete.cases(lrt2.toxb), ]
colnames(de)[colnames(de) == "logFC"] <- "log2FoldChange"
colnames(de)[colnames(de) == "PValue"] <- "pvalue"
colnames(de)[colnames(de) == "Symbol"] <- "gene_symbol"
colnames(de)[colnames(de) == "FDR"] <- "adjusted_pvalue"
de$diffabundant <- "Pass p-value cuttoff"
de$diffabundant[de$adjusted_pvalue > 0.05] <- "Not Significant"
de$diffabundant[de$log2FoldChange < -5 & de$log2FoldChange >= 5 & de$adjusted_pvalue <
0.05] <- "Pass p-value & Log2FC cuttoff"
de$diffabundant[de$log2FoldChange >= 5 & de$adjusted_pvalue <= 0.05] <- "Up"
de$diffabundant[de$log2FoldChange < -5 & de$adjusted_pvalue <= 0.05] <- "Down"
head(de) gene_symbol UID seq log2FoldChange
HGLibA_02011 ANKRD16 HGLibA_02011 CGACGTCGATGTGCCCACAC 10.154406
HGLibA_27301 LRRC29 HGLibA_27301 CTGTGTCCAGTCCCGCTTCG 7.300759
HGLibB_00250 ABHD14B HGLibB_00250 TTAAGTACCTGGCAGGTCAA 7.407535
HGLibA_42214 RPUSD4 HGLibA_42214 GACCCGAGGAATTCTCCACC 10.519154
HGLibA_39135 PSMC3IP HGLibA_39135 AATCGTGGCCCTCACTGCTA 9.567023
HGLibB_42757 SBNO2 HGLibB_42757 AGGGTATGGCGGCAGCGAGA 9.895113
logCPM LR pvalue adjusted_pvalue diffabundant
HGLibA_02011 11.875792 94.27706 2.743108e-22 1.534138e-17 Up
HGLibA_27301 11.784015 81.55213 1.706991e-19 4.773343e-15 Up
HGLibB_00250 12.149716 78.52597 7.895333e-19 1.471874e-14 Up
HGLibA_42214 10.140291 75.48743 3.677413e-18 5.141667e-14 Up
HGLibA_39135 11.552013 68.32588 1.385901e-16 1.550186e-12 Up
HGLibB_42757 9.521769 67.73967 1.865714e-16 1.739064e-12 Upg_down <- which(de$log2FoldChange > 5 & de$adjusted_pvalue <= 0.05)
g_up <- which(de$log2FoldChange < -5 & de$adjusted_pvalue <= 0.05)
de$delabel <- NA
de$delabel[c(g_down[1:20], g_up[1:20])] <- de$gene_symbol[c(g_down[1:20], g_up[1:20])]
ggplot(data = de, aes(x = log2FoldChange, y = -log10(adjusted_pvalue), col = diffabundant,
label = delabel)) + geom_point() + theme_minimal() + geom_text_repel(max.overlaps = Inf) +
scale_color_manual(values = c("blue", "black", "gray", "red")) + geom_vline(xintercept = c(-5,
5), col = "darkred") + geom_hline(yintercept = -log10(0.05), col = "darkred")