Chapter 3 g-computation
Details about standarization function in APPENDIX.
3.1 Overall
Based on Hernan and Robins, 2020 Chapter 13
f <- SEROPOSITIVE ~ work_out1 + edad + sex_male
library(boot)
# bootstrap
results <- boot(data = dat,
statistic = standardization,
treatment = work_out1, outcome = SEROPOSITIVE, formula1 = f, #parameters from function
R = 5)
# generating confidence intervals
se <- c(sd(results$t[, 1]),
sd(results$t[, 2]),
sd(results$t[, 3]),
sd(results$t[, 4]))
mean <- results$t0
ll <- mean - qnorm(0.975) * se
ul <- mean + qnorm(0.975) * se
bootstrap <-data.frame(cbind(type=c("Observed", "No Treatment", "Treatment", "Treatment - No Treatment"),
mean, se, ll, ul)) %>%
mutate_at(.vars = c(2:5), ~as.numeric(as.character(.)))
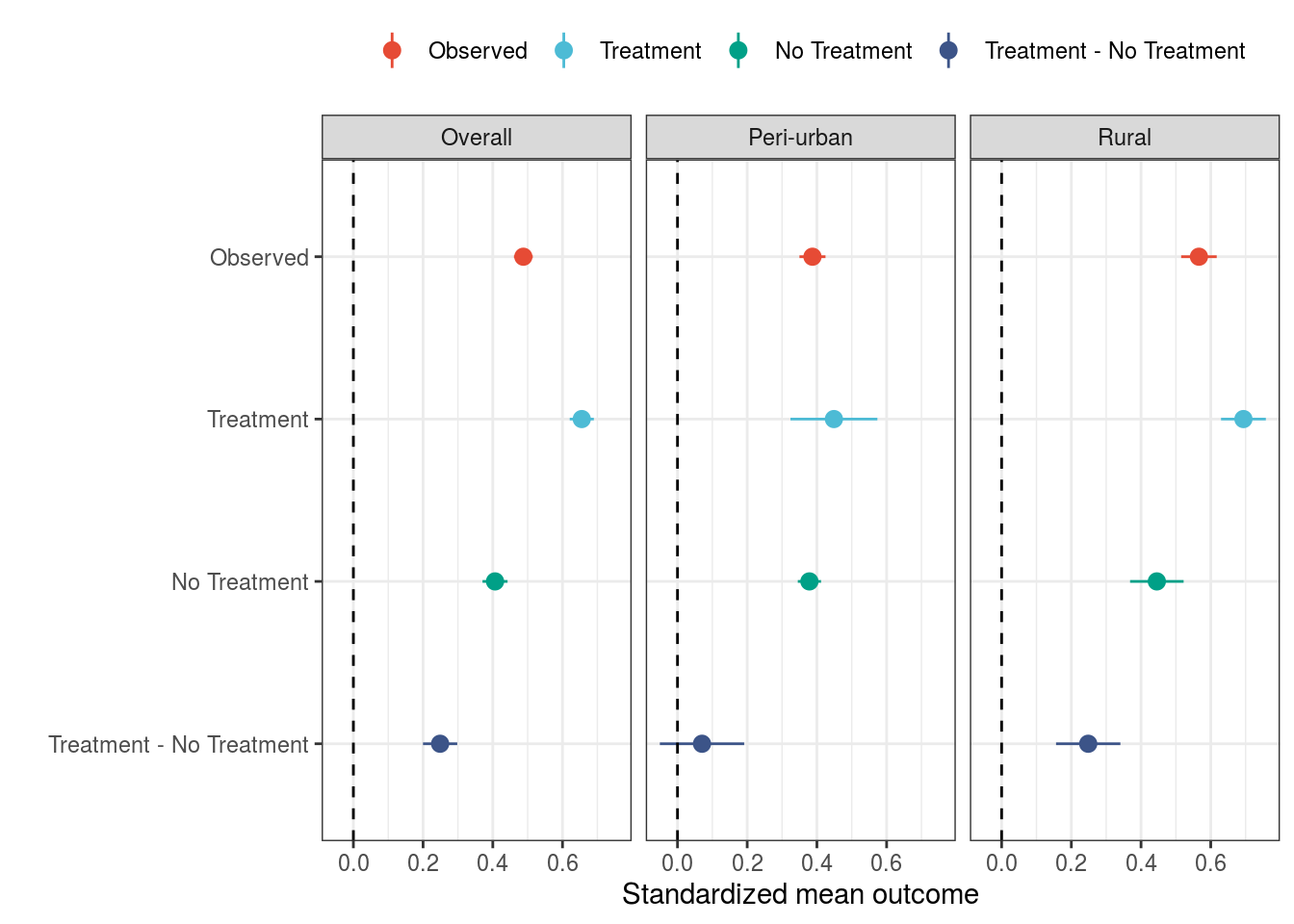
kable(bootstrap, digits = 5)| type | mean | se | ll | ul |
|---|---|---|---|---|
| Observed | 0.48771 | 0.01375 | 0.46076 | 0.51466 |
| No Treatment | 0.40645 | 0.01831 | 0.37057 | 0.44233 |
| Treatment | 0.65532 | 0.01764 | 0.62075 | 0.68989 |
| Treatment - No Treatment | 0.24887 | 0.02508 | 0.19971 | 0.29802 |
3.2 Rural
dat_rural <- dat %>%
filter(area == "1_rural")
# bootstrap
results.r <- boot(data = dat_rural,
statistic = standardization,
treatment = work_out1, outcome = SEROPOSITIVE, formula1 = f, #parameters from function
R = 5)
# generating confidence intervals
se.r <- c(sd(results.r$t[, 1]),
sd(results.r$t[, 2]),
sd(results.r$t[, 3]),
sd(results.r$t[, 4]))
mean.r <- results.r$t0
ll.r <- mean.r - qnorm(0.975) * se.r
ul.r <- mean.r + qnorm(0.975) * se.r
bootstrap.r <-data.frame(cbind(type=c("Observed", "No Treatment", "Treatment", "Treatment - No Treatment"),
mean.r, se.r, ll.r, ul.r)) %>%
mutate_at(.vars = c(2:5), ~as.numeric(as.character(.)))
kable(bootstrap.r, digits = 5)| type | mean.r | se.r | ll.r | ul.r |
|---|---|---|---|---|
| Observed | 0.56617 | 0.02602 | 0.51517 | 0.61717 |
| No Treatment | 0.44538 | 0.03918 | 0.36859 | 0.52216 |
| Treatment | 0.69384 | 0.03279 | 0.62958 | 0.75811 |
| Treatment - No Treatment | 0.24847 | 0.04710 | 0.15616 | 0.34078 |
3.3 Periurban
dat_urban <- dat %>%
filter(area == "0_periurban")
# bootstrap
results.u <- boot(data = dat_urban,
statistic = standardization,
treatment = work_out1, outcome = SEROPOSITIVE, formula1 = f, #parameters from function
R = 5)
# generating confidence intervals
se.u <- c(sd(results.u$t[, 1]),
sd(results.u$t[, 2]),
sd(results.u$t[, 3]),
sd(results.u$t[, 4]))
mean.u <- results.u$t0
ll.u <- mean.u - qnorm(0.975) * se.u
ul.u <- mean.u + qnorm(0.975) * se.u
bootstrap.u <-data.frame(cbind(type=c("Observed", "No Treatment", "Treatment", "Treatment - No Treatment"),
mean.u, se.u, ll.u, ul.u)) %>%
mutate_at(.vars = c(2:5), ~as.numeric(as.character(.)))
kable(bootstrap.u, digits = 5)| type | mean.u | se.u | ll.u | ul.u |
|---|---|---|---|---|
| Observed | 0.38726 | 0.01906 | 0.34991 | 0.42461 |
| No Treatment | 0.37848 | 0.01699 | 0.34518 | 0.41177 |
| Treatment | 0.44880 | 0.06364 | 0.32408 | 0.57353 |
| Treatment - No Treatment | 0.07033 | 0.06172 | -0.05064 | 0.19130 |
3.4 Summary g-computation
library(ggsci)
bootstrap %>% mutate(model = "Overall") %>%
rbind(bootstrap.r %>% set_names(colnames(bootstrap)) %>% mutate(model = "Rural")) %>%
rbind(bootstrap.u %>% set_names(colnames(bootstrap)) %>% mutate(model = "Peri-urban")) %>%
ggplot(aes(y = type, x = mean, col = type)) +
geom_pointrange(aes(xmin = ll, xmax = ul)) +
#geom_point() +
geom_vline(xintercept = 0, linetype = "dashed") +
scale_color_npg(limits = c("Observed", "Treatment", "No Treatment", "Treatment - No Treatment")) +
scale_y_discrete(limits = rev(c("Observed", "Treatment", "No Treatment", "Treatment - No Treatment"))) +
theme_bw() +
labs(y = "", x = "Standardized mean outcome", col = "") +
facet_grid(.~model) +
theme(legend.position = "top")