Chapter 8 FreeSurfer v6
Log onto the remote computer. Remember you will be using the preprocessed T1 image found under ‘/work/ashley.ware/data/ACAP/prepAnat/’. Make Freesurfer subjects directory.
mkdir -p /work/ashley.ware/data/ACAP/mindboggle/freesurfer_subjects/8.1 Job Script
Still on remote computer, create two job scripts. The first is for when both T1 and T2 images are available.
vi ~/scripts/ACAP/freesurferT1T2-job.shCopy and paste.
#!/bin/bash
#SBATCH --time=30:00:00 # walltime
#SBATCH --ntasks=1 # number of processor cores (i.e. tasks)
#SBATCH --nodes=1 # number of nodes
#SBATCH --mem-per-cpu=16G # memory per CPU core
# LOAD ENVIRONMENTAL VARIABLES
username=`id -un`
export FREESURFER_HOME=/home/${username}/apps/freesurfer
source $FREESURFER_HOME/SetUpFreeSurfer.sh
# INSERT CODE, AND RUN YOUR PROGRAMS HERE
recon-all \
-subjid ${1} \
-i /work/${username}/data/ACAP/prepAnat/${1}/${1}_T1w.nii.gz \
-T2 /work/${username}/data/ACAP/prepAnat/${1}/${1}_inplaneT2.nii.gz \
-T2pial \
-wsatlas \
-all \
-sd /work/${username}/data/ACAP/mindboggle/freesurfer_subjects/Create the second job script for when only T1 images are present.
vi ~/scripts/ACAP/freesurferT1-job.shCopy and paste.
#!/bin/bash
#SBATCH --time=30:00:00 # walltime
#SBATCH --ntasks=1 # number of processor cores (i.e. tasks)
#SBATCH --nodes=1 # number of nodes
#SBATCH --mem-per-cpu=16G # memory per CPU core
# LOAD ENVIRONMENTAL VARIABLES
username=`id -un`
export FREESURFER_HOME=/home/${username}/apps/freesurfer
source $FREESURFER_HOME/SetUpFreeSurfer.sh
# INSERT CODE, AND RUN YOUR PROGRAMS HERE
recon-all \
-subjid ${1} \
-i /work/${username}/data/ACAP/prepAnat/${1}/${1}_T1w.nii.gz \
-wsatlas \
-all \
-sd /work/${username}/data/ACAP/mindboggle/freesurfer_subjects/8.2 Batch Script
First check that your subjid are correct.
for i in $(find /work/ashley.ware/data/ACAP/prepAnat -name "*T1w.nii.gz"); do
subjid=`echo $i | cut -d "/" -f7`
echo $subjid
doneIf everything is correct then you can create your batch file.
Create batch script.
vi ~/scripts/ACAP/freesurfer-batch.shCopy and paste.
#!/bin/bash
curTime=`date +"%Y%m%d-%H%M%S"`
username=`id -un`
mkdir -p ~/logfiles/ACAP/${curTime}
for i in $(find /work/${username}/data/ACAP/prepAnat -name "*T1w.nii.gz"); do
subjid=`echo $i | cut -d "/" -f7`
t2image=/work/${username}/data/ACAP/prepAnat/${subjid}/${subjid}_inplaneT2.nii.gz
if [ -f $t2image ]; then
sbatch \
-o ~/logfiles/ACAP/${curTime}/output-${subjid}.txt \
-e ~/logfiles/ACAP/${curTime}/error-${subjid}.txt \
~/scripts/ACAP/freesurferT1T2-job.sh \
${subjid}
sleep 1
else
sbatch \
-o ~/logfiles/ACAP/${curTime}/output-${subjid}.txt \
-e ~/logfiles/ACAP/${curTime}/error-${subjid}.txt \
~/scripts/ACAP/freesurferT1-job.sh \
${subjid}
sleep 1
fi
done8.3 Submit Jobs with Batch Script
Submit jobs.
sh ~/scripts/ACAP/freesurfer-batch.shYou can check the status of your jobs by checking if it is running or pending.
squeueYou can check the outputs.
tail -n 5 ~/logfiles/ACAP/<date>/output*8.4 Check that FreeSurfer Completed without Errors
When FreeSurfer is finished the last line in your logfiles will tell you if FreeSurfer completed with or without errors. To check for this line specifically, use grep.
cd /home/ashley.ware/logfiles/ACAP/20190126-061608
grep -Ha "recon-all.*finished" output-sub-ACAP*You should hopefully see an output like this.
output-sub-ACAP1001_ses-BL.txt:recon-all -s sub-ACAP1001_ses-BL finished without error
output-sub-ACAP1001_ses-FU6.txt:recon-all -s sub-ACAP1001_ses-FU6 finished without error
output-sub-ACAP1002_ses-BL.txt:recon-all -s sub-ACAP1002_ses-BL finished without error
output-sub-ACAP1003_ses-BL.txt:recon-all -s sub-ACAP1003_ses-BL finished without error
output-sub-ACAP1003_ses-FU3.txt:recon-all -s sub-ACAP1003_ses-FU3 finished without error
output-sub-ACAP1004_ses-BL.txt:recon-all -s sub-ACAP1004_ses-BL finished without error
output-sub-ACAP1004_ses-FU6.txt:recon-all -s sub-ACAP1004_ses-FU6 finished without error
output-sub-ACAP1005_ses-FU3.txt:recon-all -s sub-ACAP1005_ses-FU3 finished without error
output-sub-ACAP1006_ses-BL.txt:recon-all -s sub-ACAP1006_ses-BL finished without error
output-sub-ACAP1006_ses-FU6.txt:recon-all -s sub-ACAP1006_ses-FU6 finished without error
output-sub-ACAP1009_ses-BL.txt:recon-all -s sub-ACAP1009_ses-BL finished without error8.5 Download Directory / Sync Directory with Arc
Back on your local computer make sure you have the directory already.
mkdir -p /Volumes/bobo/data/ACAP/mindboggle/freesurfer_subjectsThen sync the data from the remote computer to your local computer.
rsync -rauv \
ashley.ware@arc.ucalgary.ca:/work/ashley.ware/data/ACAP/mindboggle/freesurfer_subjects/ \
/Volumes/bobo/data/ACAP/mindboggle/freesurfer_subjects/8.6 Quality Control
Before you can run the quality control program, you’ll need to create some scripts locally on your computer. Make sure you have FreeSurfer, Homebrew, and ImageMagick installed on your local computer.
mkdir -p ~/apps/QA/
vi ~/apps/QA/takeshotsCopy and paste the following script.
#!/bin/bash
if [ -z "$1" ]; then
cat <<EOU
Capture some screenshots of surfaces produced by
FreeSurfer in standard orientations.
Usage: takeshots [options]
The options are:
-s <subjid> : Specify one subject ID
-l <listid> : Specify a file list with the subject IDs, one per line
-m <mesh> : Specify a surface file (pial, white, inflated, sphere, etc.)
-p <parc> : Specify a parcellation to load (aparc, aparc.a2009s, aparc.a2005s)
-v <volume> : Run also for volumes (0 = FALSE, 1 = TRUE)
_____________________________________
Anderson M. Winkler
Yale University / Institute of Living
Jan/2010
http://brainder.org
EOU
exit 1
fi
DOASEG="N"
# Check and accept the arguments
while getopts 's:l:m:p:v:' OPTION
do
case ${OPTION} in
s) SUBJ_LIST="${SUBJ_LIST} ${OPTARG}" ;;
l) SUBJ_LIST="${SUBJ_LIST} $(cat ${OPTARG})" ;;
m) MESH_LIST="${MESH_LIST} ${OPTARG}" ;;
p) PARC_LIST="${PARC_LIST} ${OPTARG}" ;;
v) VOLUME=${OPTARG} ;;
esac
done
for s in ${SUBJ_LIST} ; do
mkdir -p ${SUBJECTS_DIR}/${s}/shots
export SUBJECT_NAME=${s}
for h in lh rh ; do
for m in ${MESH_LIST} ; do
for p in ${PARC_LIST} ; do
export SURF=${m}
export PARC=${p}
tksurfer ${s} ${h} ${m} -tcl $(dirname $0)/shots_tksurfer.tcl
done
done
done
sleep 1
if [ ${VOLUME} == "1" ]
then
tkmedit ${s} T1.mgz -surfs -tcl $(dirname $0)/shots_tkmedit.tcl
fi
doneYou will need to create two more files.
vi ~/apps/QA/shots_tksurfer.tclCopy and paste.
# This TCL script is supposed to be called from within tksurfer, by the
# script called 'takeshots'.
# The environment variables that should be present are:
# - SUBJECTS_DIR: regular FS variable.
# - SUBJECT_NAME: name of the current subject.
# - SURF: surface to be loaded, e.g. pial, white, inflated, etc.
# - PARC: parcellation scheme, e.g. aparc, a2005s, a2009s.
#
# _____________________________________
# Anderson M. Winkler
# Yale University / Institute of Living
# Feb/2010
# http://brainder.org
open_window
set lablpth "$env(SUBJECTS_DIR)/$env(SUBJECT_NAME)/label"
set outpth "$env(SUBJECTS_DIR)/$env(SUBJECT_NAME)/shots"
labl_import_annotation "$lablpth/$hemi.$env(PARC).annot"
redraw
UpdateAndRedraw
puts "Taking Snapshots..."
make_lateral_view
redraw
set tiff "$outpth/$env(SUBJECT_NAME)_${hemi}_$env(SURF)_$env(PARC)_lat.tif"
save_tiff $tiff
rotate_brain_x 90
redraw
set tiff "$outpth/$env(SUBJECT_NAME)_${hemi}_$env(SURF)_$env(PARC)_inf.tif"
save_tiff $tiff
rotate_brain_x 180
redraw
set tiff "$outpth/$env(SUBJECT_NAME)_${hemi}_$env(SURF)_$env(PARC)_sup.tif"
save_tiff $tiff
make_lateral_view
rotate_brain_y 180
redraw
set tiff "$outpth/$env(SUBJECT_NAME)_${hemi}_$env(SURF)_$env(PARC)_med.tif"
save_tiff $tiff
exitCreate the last scripts.
vi ~/apps/QA/shots_tkmedit.tclCopy and paste.
for { set slice 0 } { $slice < 256 } { incr slice 5 } {
SetSlice $slice
RedrawScreen
set imagepath "$env(SUBJECTS_DIR)/$env(SUBJECT_NAME)/shots/$env(SUBJECT_NAME)_volume_${slice}.tif"
SaveTIFF $imagepath
}
exitNow you are ready to run takeshots which will allow you to quickly inspect FreeSurfer aseg, white matter, and cortical surfaces:
# Make output directory
mkdir -p /Volumes/bobo/data/ACAP/mindboggle/freesurfer_subjects/shots
# Set FreeSurfer subjects directory
export SUBJECTS_DIR=/Volumes/bobo/data/ACAP/mindboggle/freesurfer_subjects/
# Change into subjects directory
cd $SUBJECTS_DIR
# Get name of all subjects in FreeSurfer directory
find . -maxdepth 1 -name "sub-ACAP*" | sed 's|^./||' > subjects.txt
# Run program that will grab screenshots of results
~/apps/QA/takeshots \
-l subjects.txt \
-m pial -m inflated -p aparc -v 1Next create a montage image of the screenshots:
# Set FreeSurfer subjects directory if you haven't already
export SUBJECTS_DIR=/Volumes/bobo/data/ACAP/mindboggle/freesurfer_subjects/
# Change into subjects directory
cd $SUBJECTS_DIR
# Set output directory that you created above
OUTPUT_DIR=/Volumes/bobo/data/ACAP/mindboggle/freesurfer_subjects/shots
# Loop through subjects and combine the screenshots into a single montage image
for i in `cat subjects.txt`; do
cd $SUBJECTS_DIR/$i/shots
# Inflated Surface
montage ${i}_[lr]h_inflated* -tile 1x4 -geometry 800x800+1+1 miff:- |\
convert - +append ${OUTPUT_DIR}/${i}_inflated.png
# Pial Surface
montage ${i}_[lr]h_pial* -tile 1x4 -geometry 800x800+1+1 miff:- |\
convert - +append ${OUTPUT_DIR}/${i}_pial.png
# Tissue
montage ${i}_volume_{20..195..5}.tif -tile 6x1 -geometry 800x800+1+1 miff:- |\
convert - -append ${OUTPUT_DIR}/${i}_wm.png
doneYou can review the images afterwards. The montage images are located at /Volumes/bobo/data/ACAP/mindboggle/freesurfer_subjects/shots
The images should look like these:
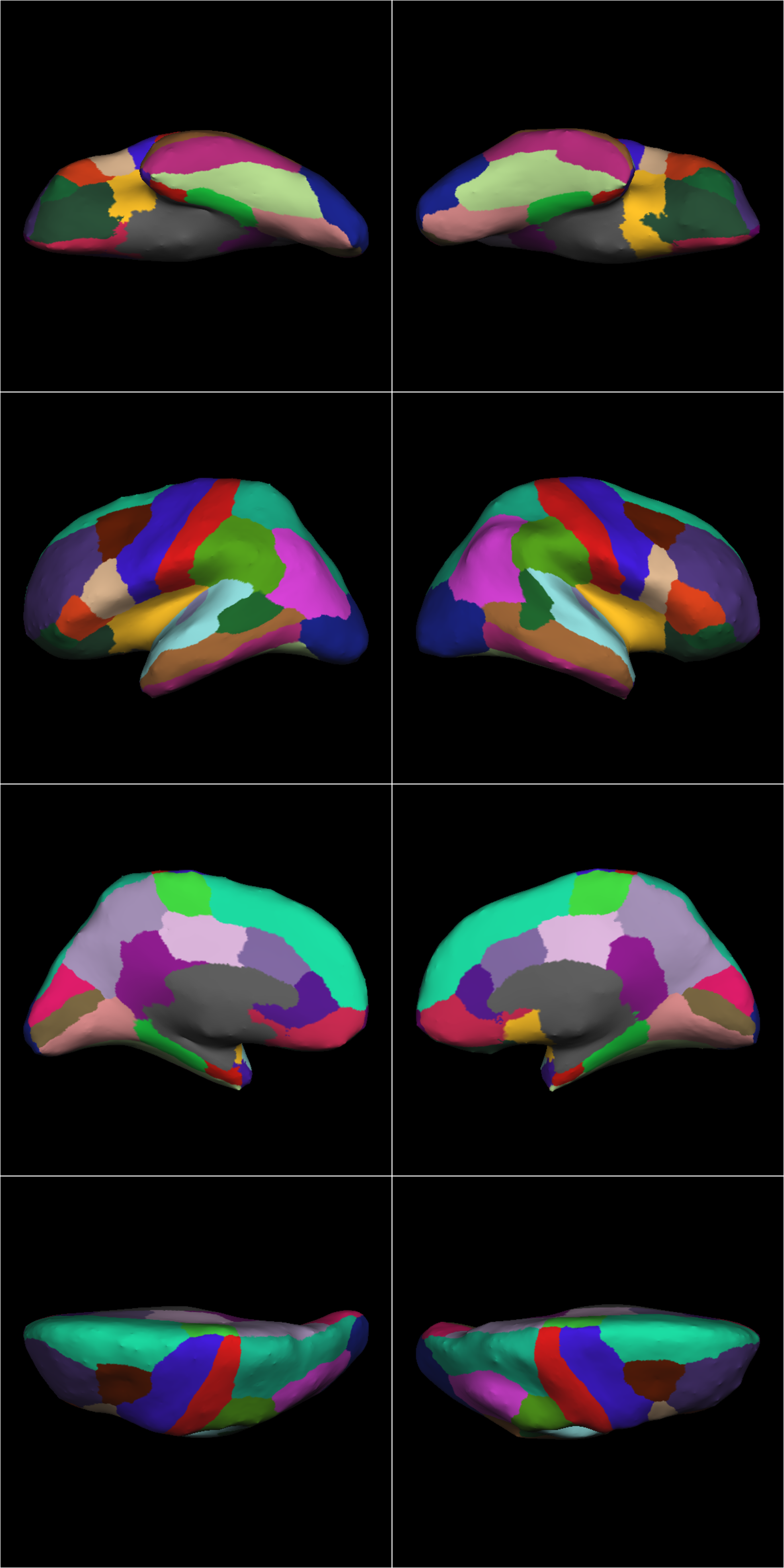
sub-ACAP1001_ses-BL_inflated
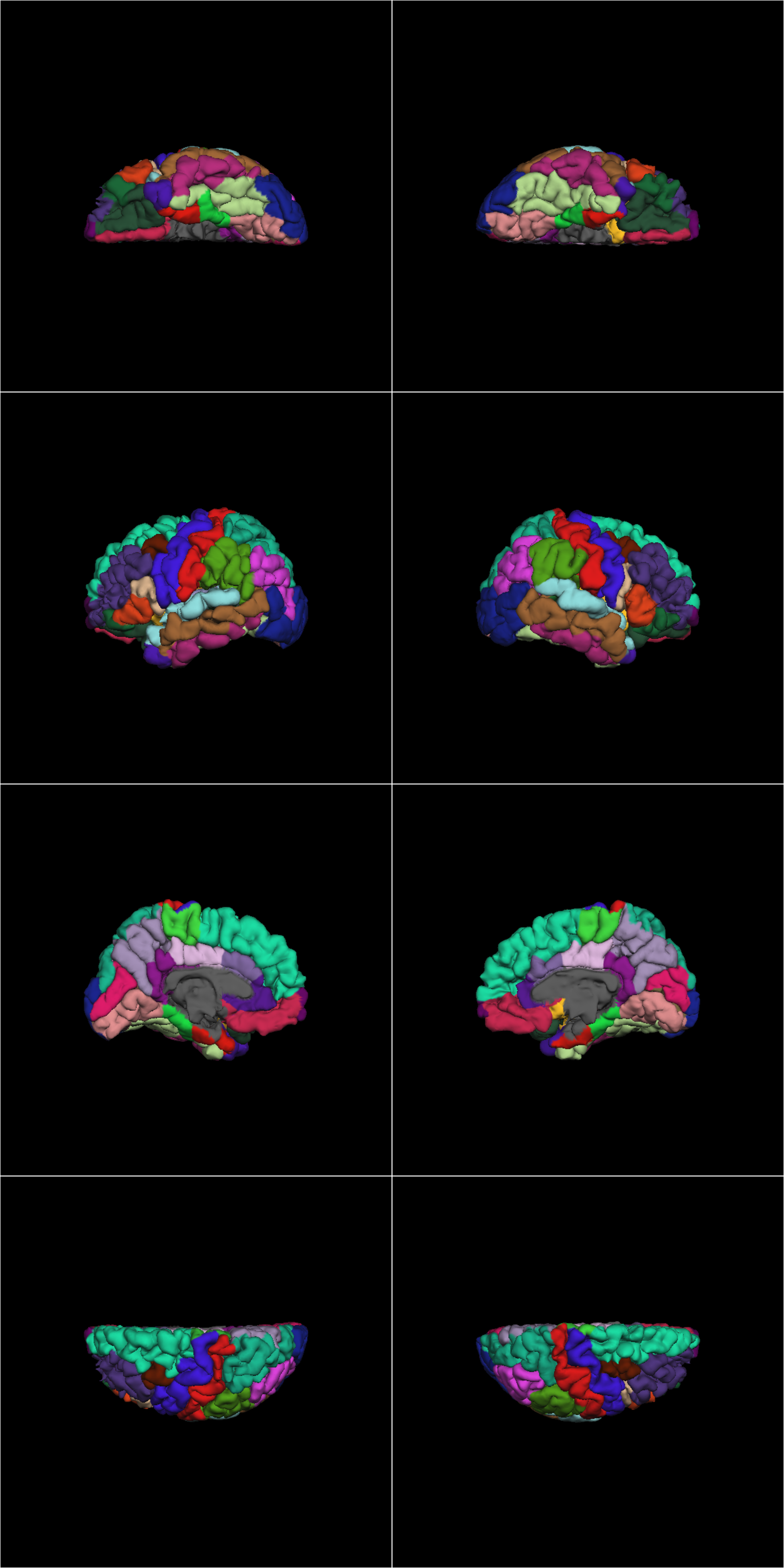
sub-ACAP1001_ses-BL_pial
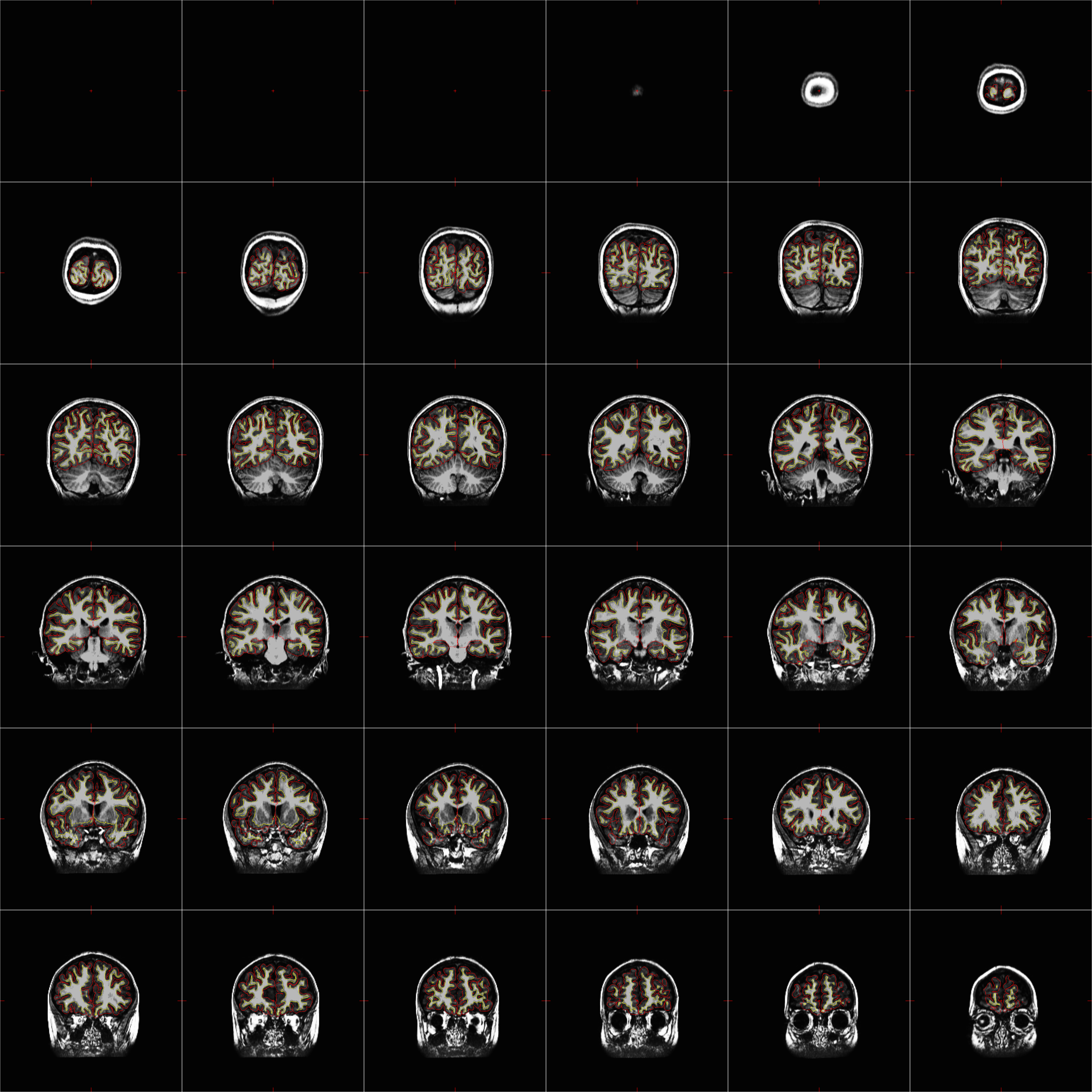
sub-ACAP1001_ses-BL_wm
8.7 Generate Tables
If there are any participants that completely fail FreeSurfer, you can remove those directory under /Volumes/bobo/data/ACAP/mindboggle/freesurfer_subjects/ and make sure to remove their name from /Volumes/bobo/data/ACAP/mindboggle/freesurfer_subjects/subjects.txt. Otherwise, any subjects in this diretory will have their data generated and placed into the output tables.
mkdir -p /Volumes/bobo/data/ACAP/mindboggle/freesurfer_subjects/tablefiles8.7.1 Cortical Parcellation
Converts a cortical stats file created by recon-all and ormris_anatomical_stats (eg, ?h.aparc.stats) into a table in which each line is a subject and each column is a parcellation. The first row is a list of the parcellation names. The first column is the subject name. By default, it looks for the ?h.aparc.stats file based on the Killiany/Desikan parcellation atlas. For more information visit https://surfer.nmr.mgh.harvard.edu/fswiki/aparcstats2table.
8.7.1.1 Area in mm2
SUBJECTS_DIR=/Volumes/bobo/data/ACAP/mindboggle/freesurfer_subjects/
cd ${SUBJECTS_DIR}
for i in lh rh; do
aparcstats2table \
--subjectsfile subjects.txt \
--hemi $i \
--meas area \
--skip \
--delimiter comma \
--tablefile tablefiles/aparc-area-${i}-stats.csv
done8.7.1.2 Cortical Thickness
SUBJECTS_DIR=/Volumes/bobo/data/ACAP/mindboggle/freesurfer_subjects/
cd ${SUBJECTS_DIR}
for i in lh rh; do
aparcstats2table \
--subjectsfile subjects.txt \
--hemi $i \
--meas thickness \
--skip \
--delimiter comma \
--tablefile tablefiles/aparc-thickness-${i}-stats.csv
done8.7.1.3 Mean Curvature
SUBJECTS_DIR=/Volumes/bobo/data/ACAP/mindboggle/freesurfer_subjects/
cd ${SUBJECTS_DIR}
for i in lh rh; do
aparcstats2table \
--subjectsfile subjects.txt \
--hemi $i \
--meas meancurv \
--skip \
--delimiter comma \
--tablefile tablefiles/aparc-meancurv-${i}-stats.csv
done8.7.2 Subcortical Segmentation
This script will generate text/ascii tables of freesurfer aseg stats data, aseg.stats. This can then be easily imported into a spreadsheet and/or stats program. For more information visit https://surfer.nmr.mgh.harvard.edu/fswiki/asegstats2table.
8.7.2.1 Volume in mm3
The hippocampus and amygdala volumes in this file are WRONG. Please verify the ROIs of any other subcortical region before using as these segmentations are often inaccurate. Refer to the seperate hippocampus / amygdala, brainstem, and thalamus pipelines.
SUBJECTS_DIR=/Volumes/bobo/data/ACAP/mindboggle/freesurfer_subjects/
cd ${SUBJECTS_DIR}
asegstats2table \
--subjectsfile subjects.txt \
--meas volume \
--skip \
--delimiter comma \
--tablefile tablefiles/aseg-volume-stats.csv