Chapter 2 Introduction to Genomics
Bioinformatics is a vast, fast-paced and complicated field of science, which focuses on using biology, computer science, mathematics and information technology to analyse biological data.
Don’t worry though, we will only focus on one key area of bioinformatics - gene expression analyses.
In order to discuss and interpret gene expression analysis results however, we will need to introduce some terms - please take a look over the following details and definitions. We hope they provide you with sufficient context to have a general understanding of the data we will be using in Computer Lab 8B.
Please note that some of the following details are slightly simplified.
2.1 DNA and Base Pairs
Since we are considering a gene expression analysis, we need to know a little about genes.
An organism’s genetic information is contained within deoxyribonucleic acid (DNA). This nucleic acid molecule is used to store huge amounts of genetic information2.
DNA is composed of four nucleotide bases (i.e. chemical building blocks):
- cytosine (C)
- guanine (G)
- adenine (A) and
- thymine (T)
In Genetics and Genomics we often talk about base pairs. What does this mean?
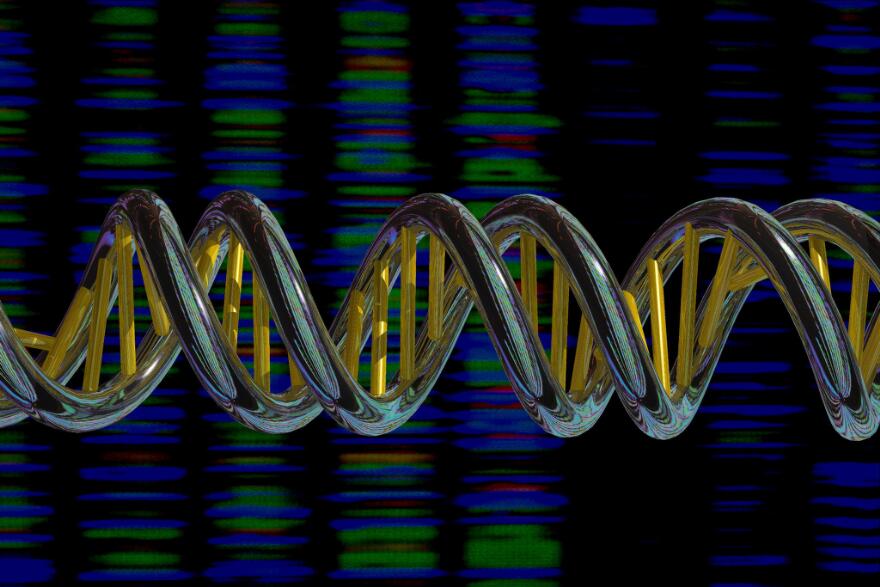
Well, let’s take a look at the image above, which is an artist’s representation of DNA. As we can see, DNA has a double-helix shape - it is double-stranded, meaning that it consists of two strands bonded together. Each strand consists of a sequence of nucleotide bases. When the strands pair to form the DNA double-helix, each nucleotide base pairs with a corresponding base on the other strand, resulting in a base pair. Due to different chemical compositions, C nucleotides can only pair with G nucleotides (resulting in a CG base pair), and A nucleotides can only pair with T nucleotides (resulting in an AT base pair).
The yellow columns in the image above represent base pairs.
To provide some context here, a human being’s total genetic information consists of around 3 billion base pairs!
2.2 Genes
A gene is a unit of genetic information, and consists of a segment of a DNA molecule. The length of a gene varies - some can be quite short while others are very long.
Genes in a eukaryotic cell (e.g. a human or plant cell) are contained within a structure made of a complete DNA molecule, which is known as a chromosome3. Humans, for example, have 23 different chromosomes.
The total genetic information that comprises an organism is referred to as the organism’s genome4.
It’s worth noting here that the overall size of an organism’s genome is not necessarily indicative of the complexity of an organism. An organism with more base pairs than another organism is not necessarily a ‘superior’ organism. For example, the wheat genome is roughly 5 times larger than the human genome!5
2.3 Gene Expression
Gene expression is a vital biological process whereby information from a gene is used in the creation of proteins and other requisite gene products6. While a change in the expression status of one gene might not do much (a protein may briefly stop/start being created), the complex interplay of simultaneous changes in gene expressions within an organism can result in significant changes to an organism over time.
For example, we tend to grow taller as we age from children to adults, and to a certain extent this is due to changes in gene expression over this period.
Gene expression is a natural process, and it is normal for the expression status of a gene to change over time. However, sometimes a gene may be negatively influenced (by various factors), such that normal gene expression is inhibited. This can have serious consequences for an organism - for example, in humans, aberrant gene expression can contribute to neurological disorders and the development of certain diseases.
In Bioinformatics, we are often interested in conducting gene expression analyses, where we compare changes in the expression status of genes between individuals or groups exhibiting different characteristics. For example we may compare gene expression between cells from a healthy individual and cells from an individual with a certain disease, in order to determine potentially beneficial medical treatments.
If there is no great change in the expression status of a gene between the two groups under consideration, then that gene is probably not contributing to the differences exhibited by the two groups.
If however the change in gene expression is large for a particular gene, we say that the gene is differentially expressed. We can then use statistical tests to determine whether this differential expression is statistically significant.
2.4 RNA
When we analyse gene expression data, we actually analyse ribonucleic acid (RNA) data.
During gene expression, information from a gene is copied from DNA to RNA (transcription), and these working copies are then used to create proteins and other requisite gene products (translation)7.