Chapter 2 Introduction to Bioinformatics
Bioinformatics is a vast, fast-paced and complicated field of science, which focuses on using biology, computer science, mathematics and information technology to analyse biological data.
Don’t worry though, we will only focus on one key area of bioinformatics - gene expression analyses.
In order to conduct a small gene expression analysis however, we will need to introduce some terms - please take a look over the following details and definitions. We hope they provide you with sufficient context to have a general understanding of the data we will be using in Computer Lab 8B.
Please note that some of the following details are slightly simplified.
2.1 DNA and Base Pairs
Since we are conducting a gene expression analysis, we need to know a little about genes.
An organism’s genetic information is contained within deoxyribonucleic acid (DNA). This nucleic acid molecule is used to store huge amounts of genetic information2.
DNA is composed of four nucleotide bases (i.e. chemical building blocks):
- cytosine (C)
- guanine (G)
- adenine (A) and
- thymine (T)
In Bioinformatics we often talk about base pairs. What does this mean?
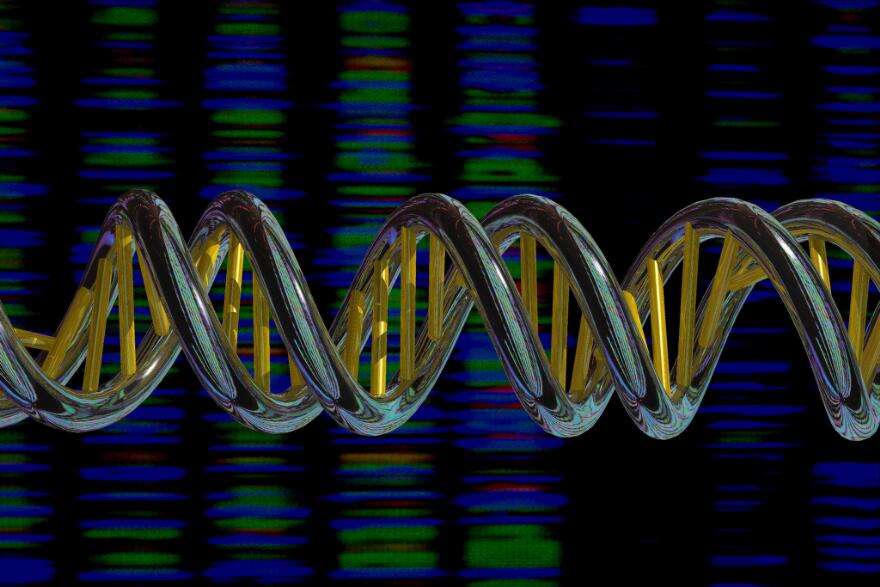
Well, let’s take a look at the image above, which is an artist’s representation of DNA. As we can see, DNA has a double-helix shape - it is double-stranded, meaning that it consists of two strands bonded together. Each strand consists of a sequence of nucleotide bases. When the strands pair to form the DNA double-helix, each nucleotide base pairs with a corresponding base on the other strand, resulting in a base pair. Due to different chemical compositions, C nucleotides can only pair with G nucleotides (resulting in a CG base pair), and A nucleotides can only pair with T nucleotides (resulting in an AT base pair).
The yellow columns in the image above represent base pairs.
To provide some context here, a human being’s total genetic information consists of around 3 billion base pairs!
2.2 Genes
A gene is a unit of genetic information, and consists of a segment of a DNA molecule. The length of a gene varies - some can be quite short while others are very long.
Genes in a eukaryotic cell (e.g. a human or plant cell) are contained within a structure made of a complete DNA molecule, which is known as a chromosome3. Humans, for example, have 23 different chromosomes.
The total genetic information that comprises an organism is referred to as the organism’s genome4.
It’s worth noting here that the overall size of an organism’s genome is not necessarily indicative of the complexity of an organism. An organism with more base pairs than another organism is not necessarily a ‘superior’ organism. For example, the wheat genome is roughly 5 times larger than the human genome!5
2.3 Gene Expression
A collection of chemical compounds and proteins known as the epigenome encompasses the genome of an organism.
If you think of an organism’s genome as a car, the epigenome is the driver.
In other words, the epigenome exerts control over the function of DNA and the production of protein in an organism6.
In particular, the epigenome controls gene expression, a vital biological process whereby information from the gene is used in the creation of proteins and other requisite gene products7. While a change in the expression status of one gene might not do much (a protein may briefly stop/start being created), the complex interplay of simultaneous changes in gene expressions within an organism can result in significant changes to an organism over time. For example, we tend to grow taller as we age from children to adults, and to a certain extent this is due to changes in gene expression over this period.
Gene expression is a natural process, and it is normal for the expression status of a gene to change over time. However, sometimes a gene may be negatively influenced (by various factors), such that normal gene expression is inhibited. This can have serious consequences for an organism - for example, in humans, aberrant gene expression can contribute to neurological disorders and the development of certain diseases.
2.4 RNA
When we analyse gene expression data, we actually analyse ribonucleic acid (RNA) data.
During gene expression, information from a gene is copied from DNA to RNA (transcription), and these working copies are then used to create proteins and other requisite gene products (translation)8.
2.5 Read counts
When we sequence an organism’s DNA or RNA, we are effectively applying a set of procedures to the DNA/RNA, in order to obtain a genetic blueprint for that organism.
We don’t need to go into the full details about the different sequencing methods available - for the time being it is sufficient to know that RNA-Seq data is obtained by sequencing and aligning randomly divided chunks of genetic material from an organism to create an overall genetic blueprint - you can think of this as being a bit like taking an artwork, turning it into a jigsaw, and then connecting all the pieces.
The sequenced RNA is referred to as the RNA library.
The number of chunks that overlap a genomic region containing a gene is referred to as the read count for that gene. The library size denotes the total number of read counts (for all genes) produced via the sequencing.
To ensure that the blueprint is accurate, these random chunks are quite small (often 300 base pairs or less)9. This means that there are plenty of chunks that overlap each other. As a result, gene read counts are often quite high - which is a good thing, as it means we can be confident in the accuracy of the sequencing process.
2.6 Sequencing Depth
The number of read counts for a gene can be referred to as the sequencing depth or sequencing coverage for that gene.
Because the sequencing process is to a certain extent random (plus we have potential for human error), there will be some unavoidable sampling variation between genes - some genes will have a lower read count than others.
To help mitigate this, multiple samples are typically used when conducting a sequencing process. This results in data for several biological replicates.
2.7 Biological Variation
Biological variation is the coefficient of variation component that would remain between biological replicates even if the sequencing depths for the samples could be increased indefinitely10.
References
“DNA double helix and sequencing output” is licensed under CC BY 4.0↩︎
Clark and Pazdernik (2013), pp.41-42↩︎
Clark and Pazdernik (2013), p. 6↩︎
Clark and Pazdernik (2013), p.38↩︎
Clark and Pazdernik (2013), p.9↩︎
McCarthy, Chen, and Smyth (2012), p.4291↩︎