4 Fig 2
4.1 Fig 2C
knitr::opts_chunk$set(warning=FALSE, message=FALSE)nair_pc1 = read.table('data/hNAD_PC1')
b_compartment = read.table('data/Other_PC1')
df = data.frame(pc1 = c(nair_pc1$V6,b_compartment$V6),group = c(rep('hNAD',dim(nair_pc1)[1]),
rep('non-hNAD',dim(b_compartment)[1])),
dose = 'B compartment')
library(ggplot2)
bp <- ggplot(df, aes(x=dose, y=pc1, fill=group)) +
geom_boxplot(width=0.4)+scale_fill_manual(values=c("#2BA809", "#00BFC4"))+
labs(title="",x="", y = "PC1")+ylim(-0.05,0)
bp + theme_classic()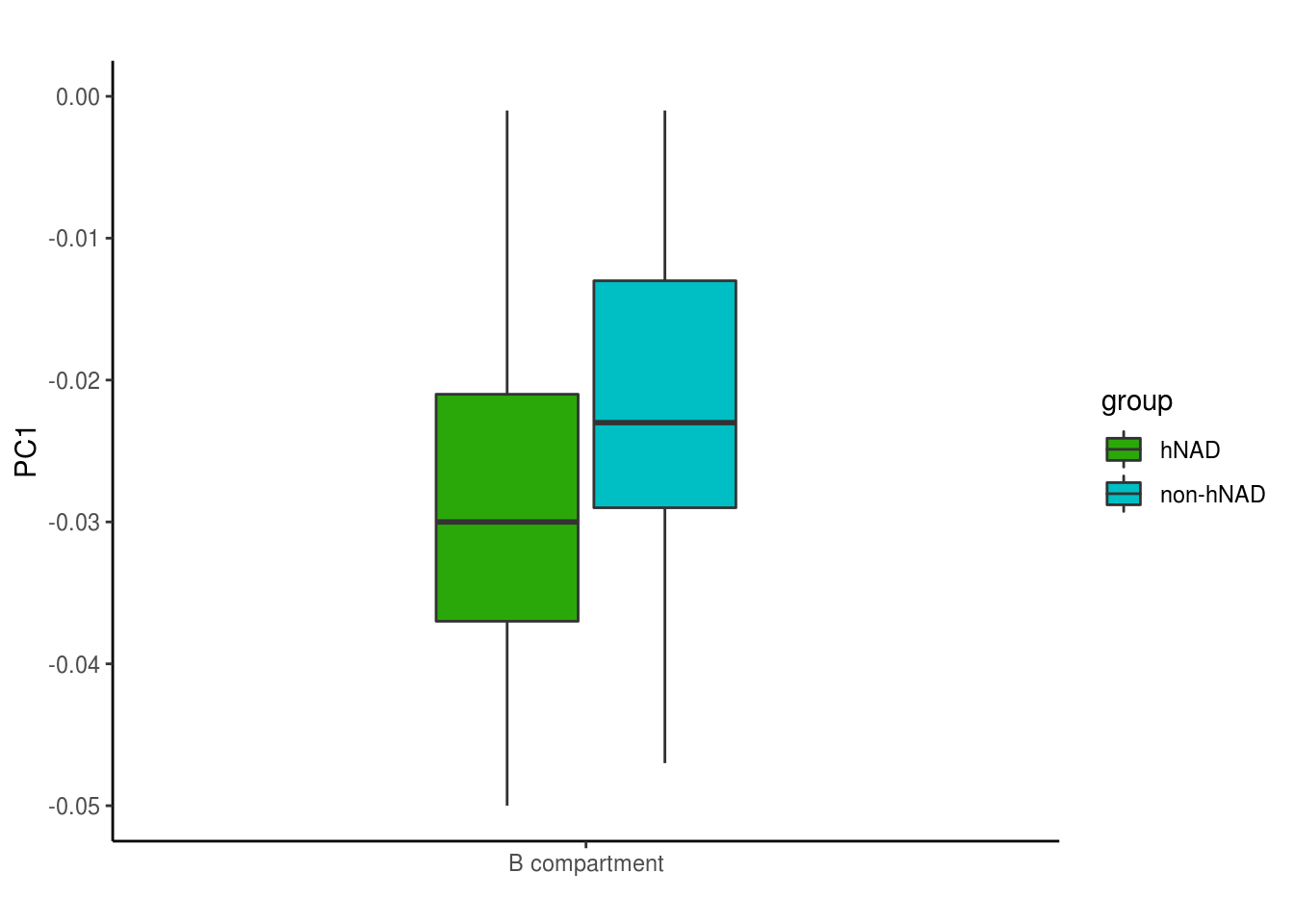
4.2 Fig 2D
hNAD = read.table('data/hNAD',stringsAsFactors = F)
hNAD$distance = 0
acen = read.table('data/acen.bed',stringsAsFactors = F)
for(i in 1:dim(hNAD)[1]){
chr = hNAD[i,1]
t = acen[acen$V1==chr,]
hNAD[i,4] = min(abs(hNAD[i,2]-t[1,2]),abs(hNAD[i,2]-t[1,3]),
abs(hNAD[i,3]-t[1,2]),abs(hNAD[i,3]-t[1,3]))
}
B = read.table('data/B_compartment.bed',stringsAsFactors = F)
B = B[,1:3]
B$distance = 0
for(i in 1:dim(B)[1]){
chr = B[i,1]
t = acen[acen$V1==chr,]
B[i,4] = min(abs(B[i,2]-t[1,2]),abs(B[i,2]-t[1,3]),
abs(B[i,3]-t[1,2]),abs(B[i,3]-t[1,3]))
}
# Density estimations
denx <- density(hNAD$distance)
deny <- density(B$distance)
# Plot
plot(denx,
ylim = c(0, max(c(denx$y, deny$y))),
xlim = c(min(c(denx$x, deny$x)),
max(c(denx$x, deny$x))),main='')
lines(deny)
polygon(denx, col = rgb(43/255, 168/255, 9/255, alpha = 0.6))
polygon(deny, col = rgb(16/255, 137/255, 114/255, alpha = 0.6))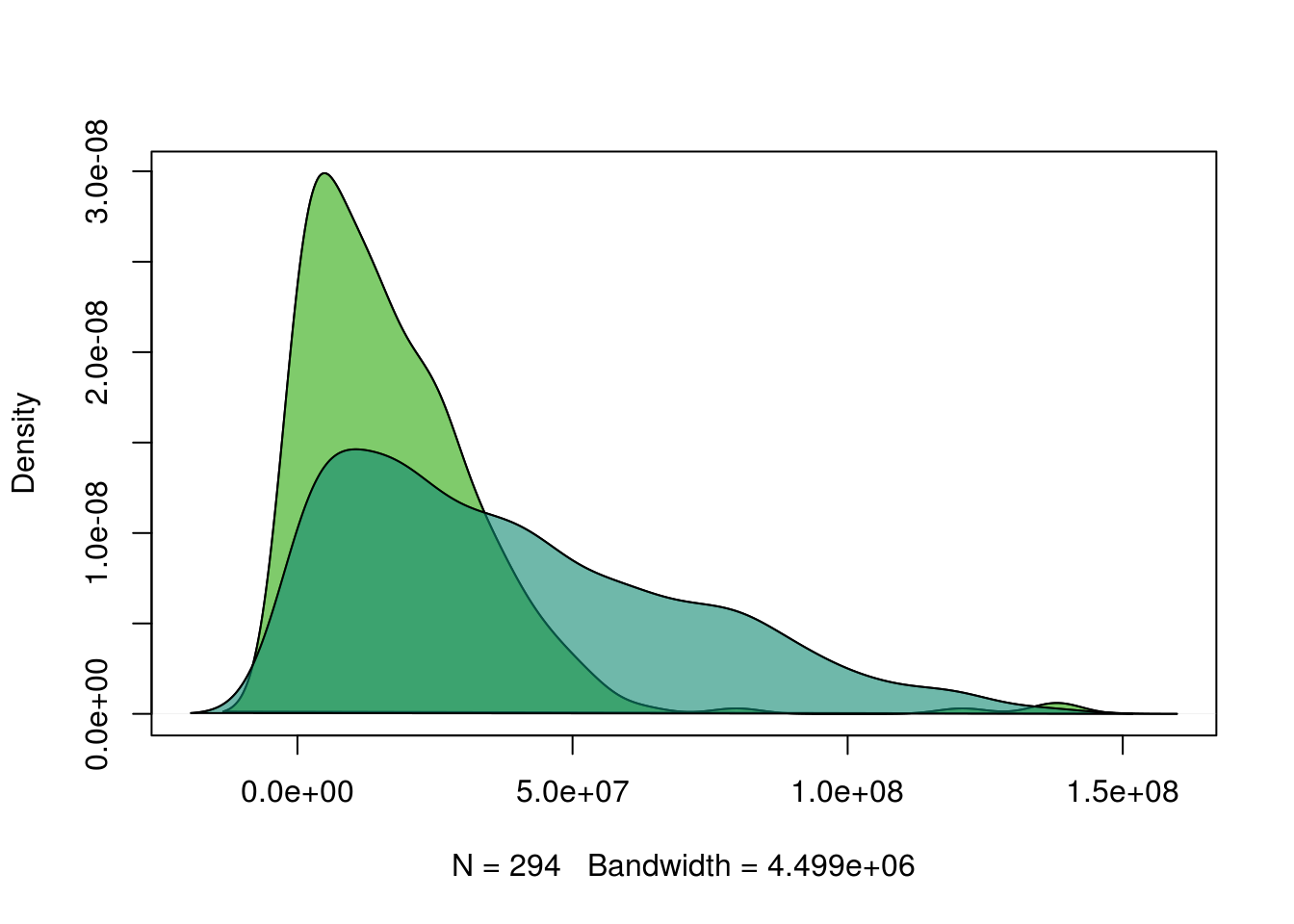
4.3 Fig 2E
nad = read.table('data/hNAD_B_gene_num')
inad = read.table('data/other_B_gene_num')
nad = nad[,4]/(nad[,3]-nad[,2])*1000000
inad = inad[,4]/(inad[,3]-inad[,2])*1000000
df = data.frame(variable=c(rep('hNAD',length(nad)),rep('non-hNAD',length(inad))),value=c(nad,inad))
ggplot(df, aes(variable, value))+
geom_boxplot(width = 0.4, notchwidth = 1, outlier.shape = NA,aes(fill=variable)) +
labs(x=NULL,y="Gene count/Mb") +
theme_classic() +
theme(text = element_text(size = 20),legend.position = "none") +
scale_fill_manual(values=c( "#2BA809","#00BFC4"))+ylim(0,25) 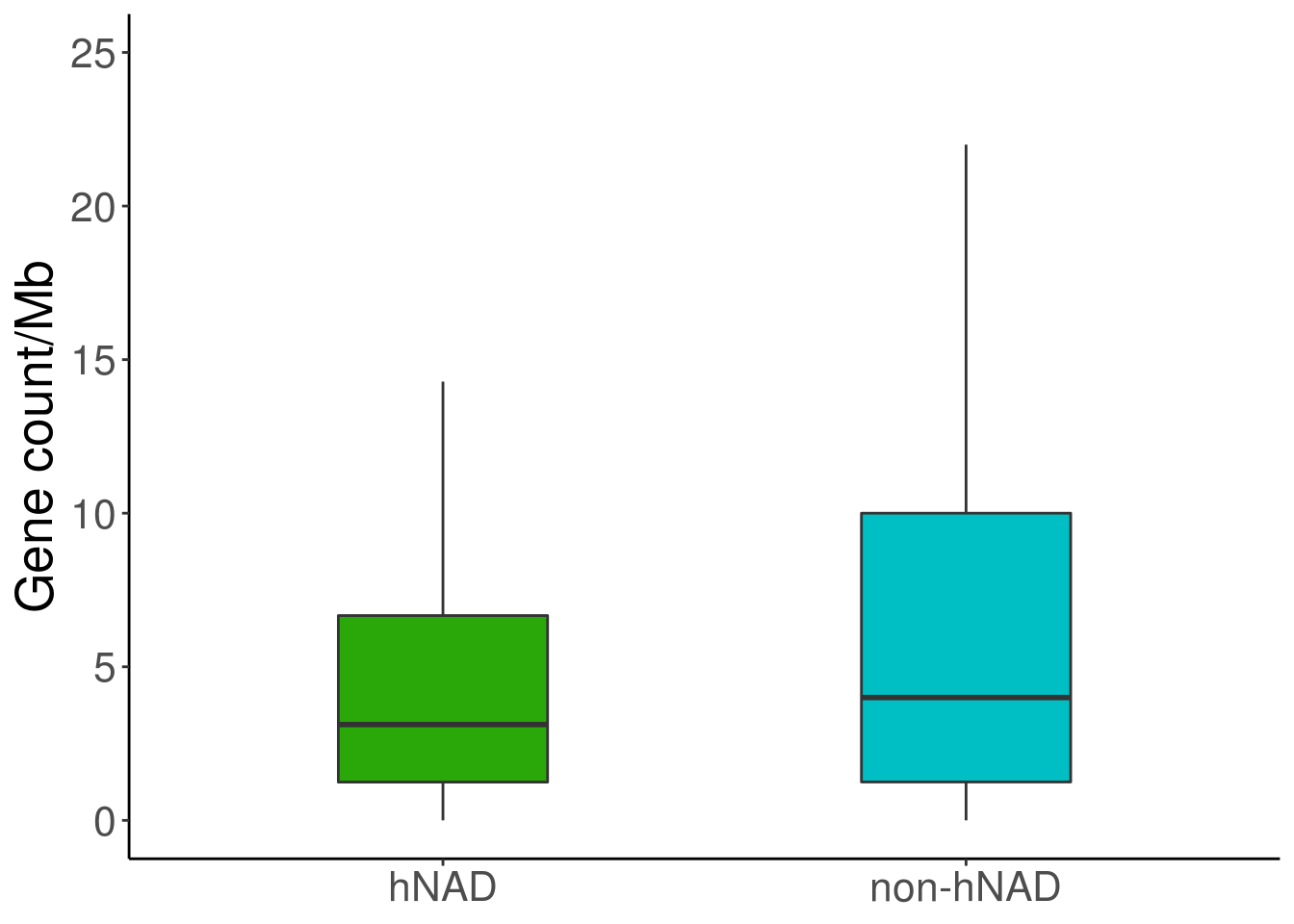
4.4 Fig 2G and 2H
bb = read.table('data/nonhNAD_participated_BB_interaction')
bb = bb[bb$V1!=bb$V4,]
hnadb = read.table('data/hNAD_participated_BB_interaction')
hnadb = hnadb[hnadb$V1!=hnadb$V4,]
chr_inter = data.frame(chr=paste0('chr',c(2:22)),hNAD=0,non_hNAD=0)
for(i in 2:22){
t = sum(bb[bb$V1==paste0('chr',i),7])
chr_inter[chr_inter$chr==paste0('chr',i),3] = t
tt = sum(hnadb[hnadb$V1==paste0('chr',i),7])
chr_inter[chr_inter$chr==paste0('chr',i),2] = tt
}
df = data.frame(inter = c(chr_inter$hNAD,chr_inter$non_hNAD),group = c(rep('hNAD',21),
rep('non-hNAD',21)),
dose = 'B compartment')
suppressMessages(library(ggplot2))
bp <- ggplot(df, aes(x=dose, y=inter, fill=group)) +
geom_boxplot(width=0.4)+
scale_fill_manual(values=c( "#2BA809","#00BFC4"))+
labs(title="",x="", y = "Normalized interaction")+ylim(0,1000000)
bp + theme_classic()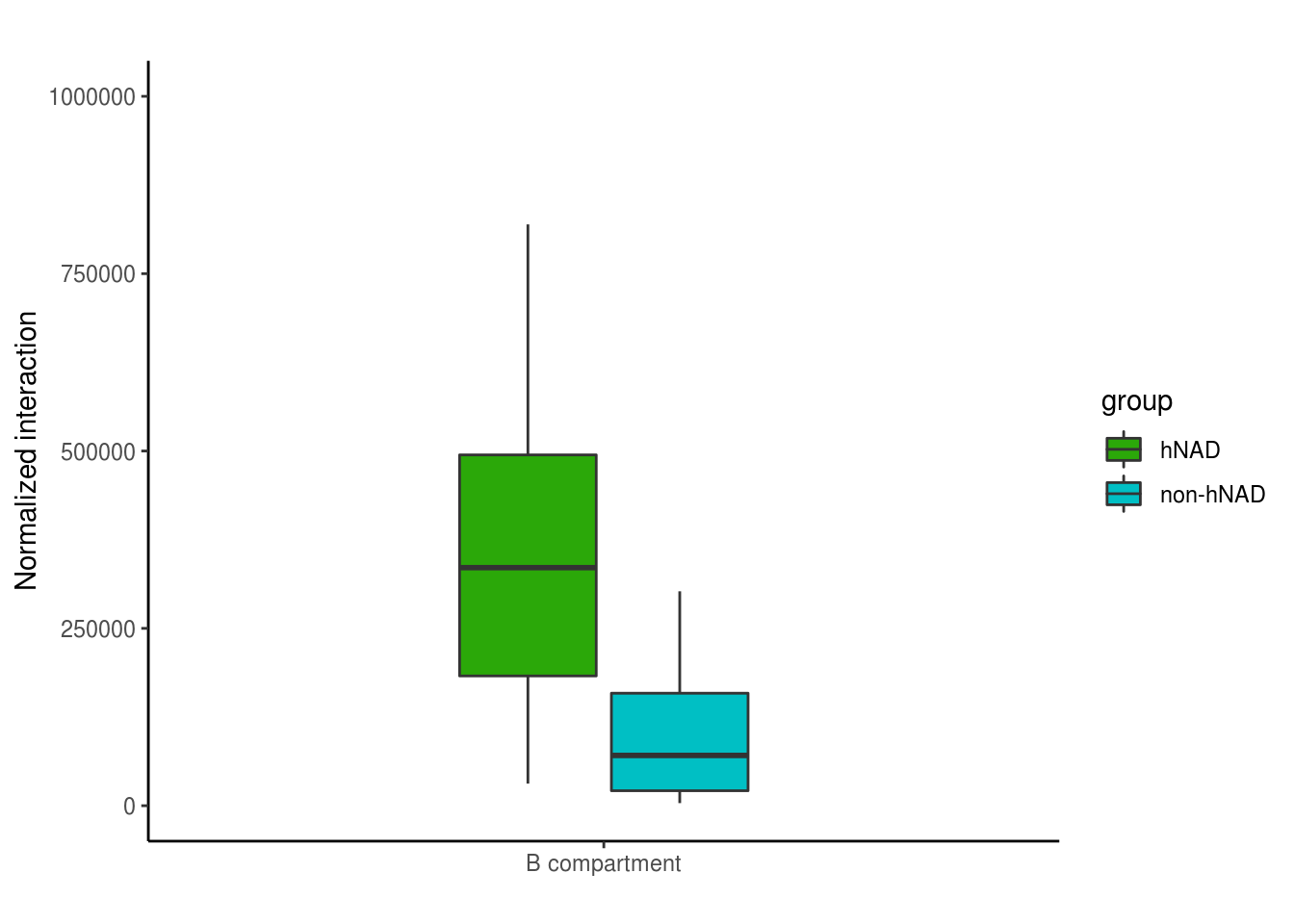
bb = read.table('data/BB_interaction')
hnadb = read.table('data/hNAD_participated_BB_interaction')
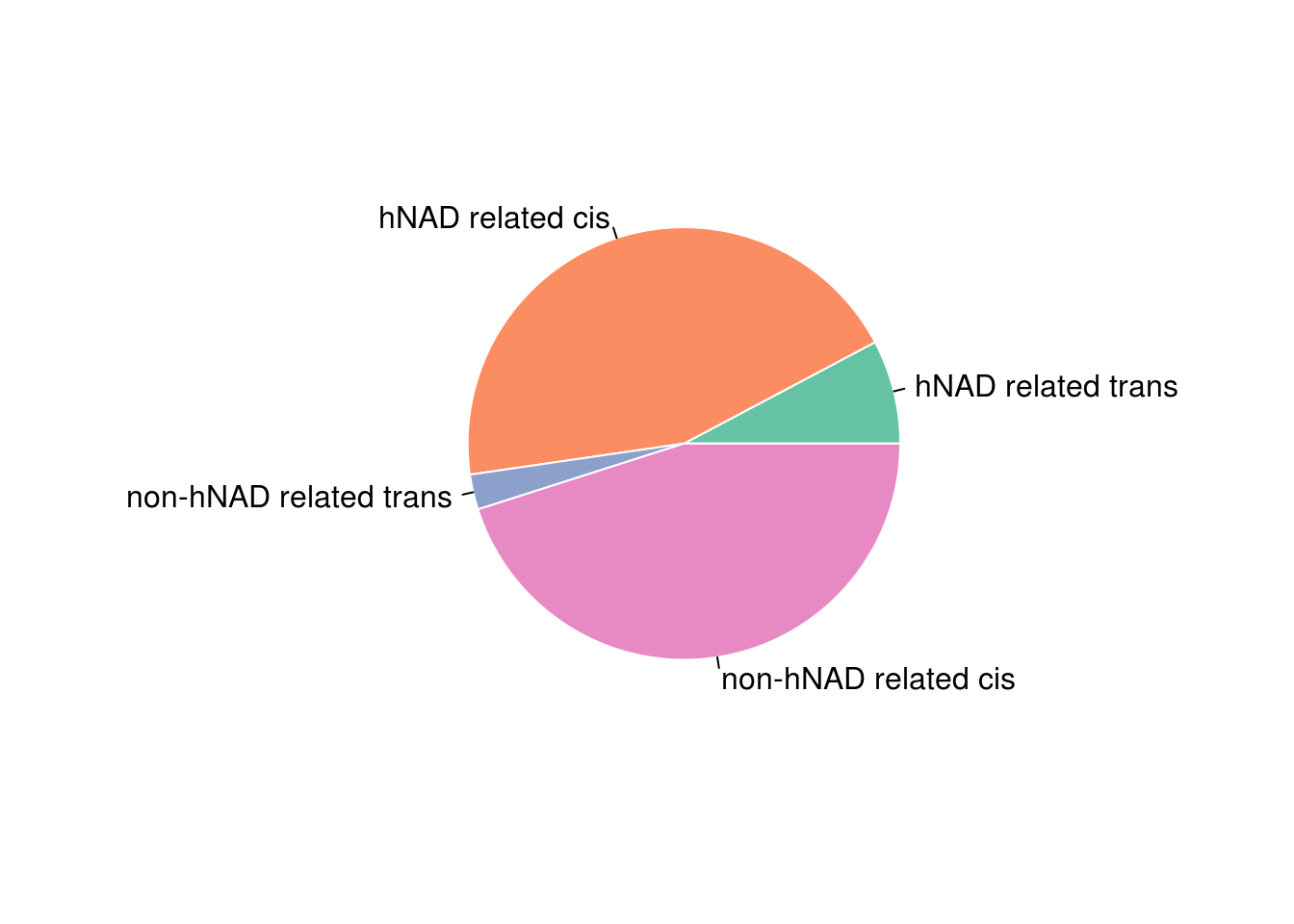
Prop = c(sum(hnadb[hnadb$V1!=hnadb$V4,7]),sum(hnadb[hnadb$V1==hnadb$V4,7]),
sum(bb[bb$V1!=bb$V4,7])-sum(hnadb[hnadb$V1!=hnadb$V4,7]),
sum(bb[bb$V1==bb$V4,7])-sum(hnadb[hnadb$V1==hnadb$V4,7]))/sum(bb$V7)
library(RColorBrewer)
myPalette <- brewer.pal(6, "Set2")
pie(Prop , labels = c('hNAD related trans','hNAD related cis','non-hNAD related trans','non-hNAD related cis'), border="white", col=myPalette )