Chapter 3 Coding Practices
3.1 Efficient Coding
This section explains how to make code run faster.1
The most basic practice in efficient coding is to keep your R, RStudio, and package versions up to date. Check you version from the version global list object.
version## _
## platform x86_64-w64-mingw32
## arch x86_64
## os mingw32
## system x86_64, mingw32
## status
## major 4
## minor 1.0
## year 2021
## month 05
## day 18
## svn rev 80317
## language R
## version.string R version 4.1.0 (2021-05-18)
## nickname Camp PontanezenUpdate R from inside the R GUI.
installr::updateR()Update RStudio from Help > Check for Updates. RStudio closes for the update. Once updated, RStudio should default to using the new version of R too.
Update your packages from the RStudio’s Packages panel.
3.1.1 Benchmarking
Benchmarking is the capture of the performance time for comparison to alternative solutions. Benchmark a section of code by wrapping it within a function and calling the function with system.time(). Ignore the user and system times - they are components of the overall elapsed time.
my_f <- function(n) {
for(i in 1:n) { x <- runif(1) }
}
system.time(my_f(1e4))## user system elapsed
## 0.03 0.00 0.03The benchmark() function in the microbenchmark package does this, but also compares functions, runs them multiple times, and calculates summary statistics.
library(microbenchmark)
microbenchmark(my_f(1e3), my_f(1e4), my_f(1e5), times = 10)## Unit: milliseconds
## expr min lq mean median uq max neval cld
## my_f(1000) 1.0607 1.0897 1.15268 1.13025 1.1679 1.3462 10 a
## my_f(10000) 11.5730 12.1453 13.60281 12.88000 15.1785 16.0651 10 b
## my_f(1e+05) 127.9111 135.7143 137.79381 136.74075 140.6062 145.7768 10 cThe benchmark_io() function in the benchmarkme package reads and writes a file and compares your performance to other users.
library(benchmarkme)
# read/write a 5MB file
my_io <- benchmark_io(runs = 1, size = 5)
## Preparing read/write io
## # IO benchmarks (2 tests) for size 5 MB:
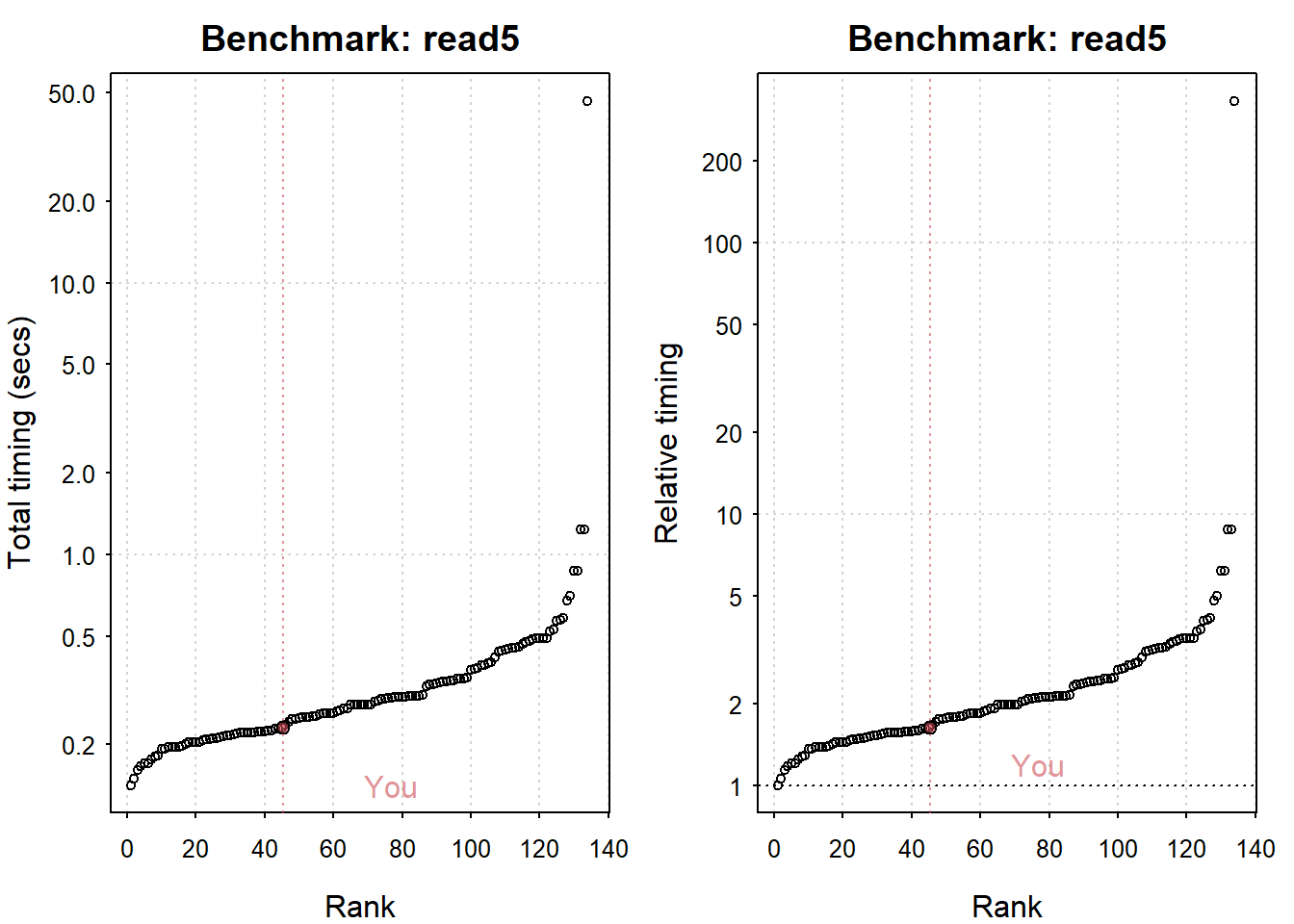
## Writing a csv with 625000 values: 1.5 (sec).
## Reading a csv with 625000 values: 0.23 (sec).
plot(my_io)
## You are ranked 79 out of 135 machines.
## Press return to get next plot
## You are ranked 46 out of 135 machines.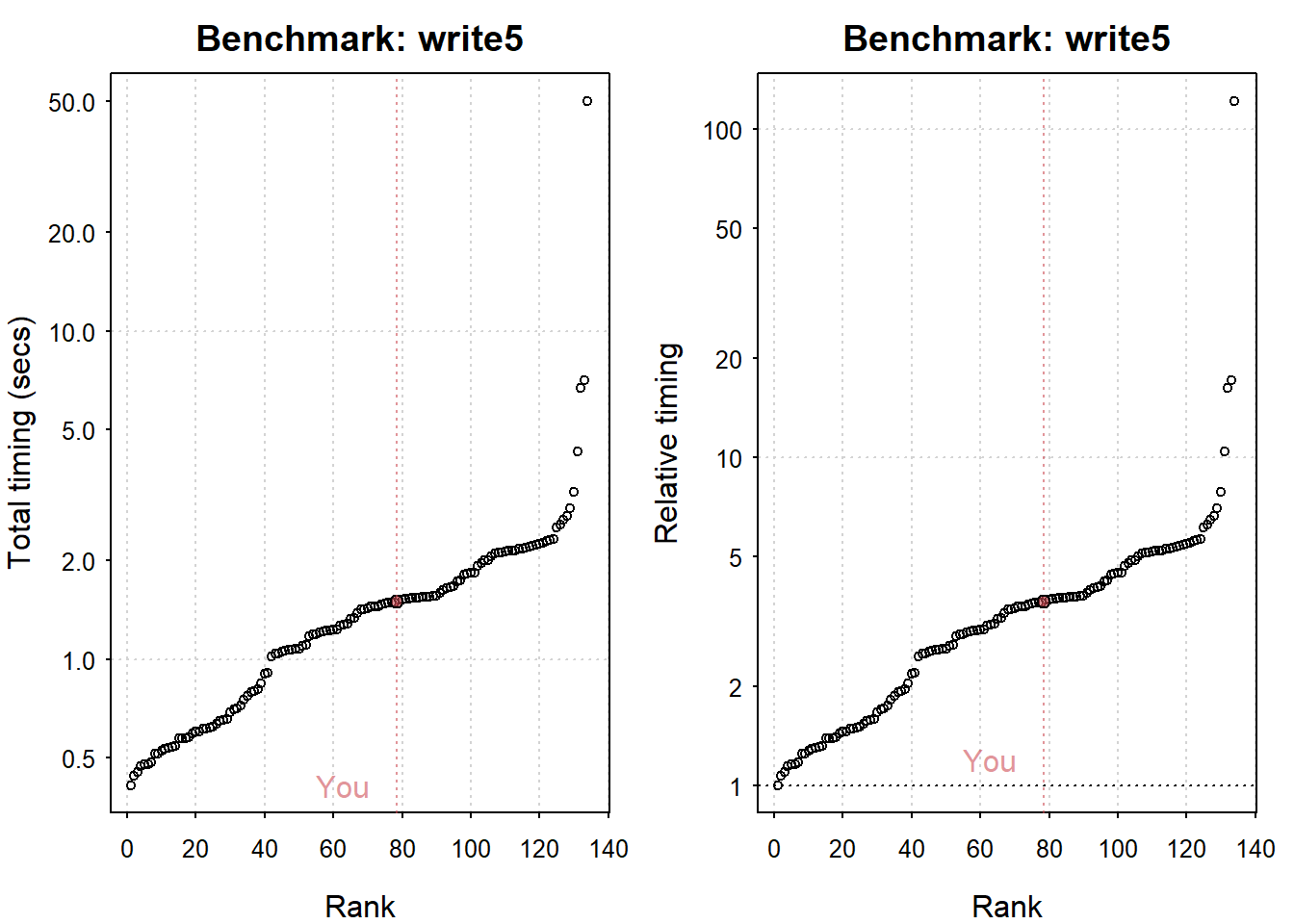
You can also use the package to retrieve hardware data.
get_ram()
## 8.26 GB
get_cpu()
## $vendor_id
## [1] "GenuineIntel"
##
## $model_name
## [1] "Intel(R) Core(TM) i5-1035G1 CPU @ 1.00GHz"
##
## $no_of_cores
## [1] 83.1.2 Profiling
Code profiling is taking time snapshots at intervals throughout the code in order to find the location of bottlenecks. Base R function Rprof() does this, but it is not user friendly. Instead, use profvis() from the profvis package.
library(profvis)
profvis({
for(i in 2:3) {
my_f(10^i)
}
my_f(1e4)
my_f(1e5)
})3.1.3 Parallel Programming
Use the parallel package to parallelize your code. Parallelization adds communication overhead among the cpus, so it’s not always helpful.
library("parallel")
mat <- as.matrix(mtcars)
# make a cluster using all cores, or maybe all but one
n_cores <- benchmarkme::get_cpu() %>% pluck("no_of_cores") - 1
cl <- makeCluster(n_cores)
# make copies of data and functions for each cluster
clusterExport(cl, "my_f")
system.time(my_f(1e5))
## user system elapsed
## 0.15 0.00 0.16
# use a parallel version of a function, like parApply instead of apply.
# In this case, the serial version is faster!
microbenchmark(apply(mat, 1, median),
parApply(cl, mat, 1, median),
times = 100)
## Unit: microseconds
## expr min lq mean median uq max
## apply(mat, 1, median) 565.6 661.5 727.009 722.70 795.45 1028.9
## parApply(cl, mat, 1, median) 1298.9 1433.5 1583.309 1542.65 1664.95 3264.8
## neval cld
## 100 a
## 100 b
# stop the cluster
stopCluster(cl)3.1.4 Other Efficiency Tips
Tip #1: Don’t allocate memory on the fly.
# bad
fun_bad <- function(n) {
x <- NULL
for(i in 1:n) { x <- c(x, rnorm(1)) }
}
# good
fun_good <- function(n) {
x <- numeric(n)
for(i in 1:length(x)) { x[i] <- rnorm(1) }
}
microbenchmark(fun_bad(1000), fun_good(1000), times = 10)
## Unit: milliseconds
## expr min lq mean median uq max neval cld
## fun_bad(1000) 1.8543 1.8996 3.30695 1.92425 4.9103 8.1975 10 a
## fun_good(1000) 1.1388 1.1671 1.63411 1.28885 1.4232 4.8641 10 aTip #2: Use a vectorized solution whenever possible.
# makes 100 calls to rnorm() and makes 100 assignments to x
x <- numeric(100)
for(i in 1:length(x)) { x[i] <- rnorm(1) }
# makes 1 call to rnorm() and 1 assignment to x
x <- rnorm(100)Tip #3: Use a matrix instead of a dataframe if possible. Matrix operations are fast because with predefined dimensions, accessing any row, col, or cell is a multiple of a dimension length.
# matrix is faster for column selection...
mat <- mtcars %>% as.matrix()
df <- mtcars
microbenchmark(mat[, 1], df[, 1])
## Unit: nanoseconds
## expr min lq mean median uq max neval cld
## mat[, 1] 800 1000 1190 1100 1200 8100 100 a
## df[, 1] 4700 5000 5648 5150 5300 50400 100 b
# and even faster for row selection (because of variable data types.
microbenchmark(mat[1, ], df[1, ])
## Unit: nanoseconds
## expr min lq mean median uq max neval cld
## mat[1, ] 500 600 979 800 1000 6900 100 a
## df[1, ] 53500 55400 64825 56000 61000 151800 100 bTip #4: Use && for smarter logical testing - if condition 1 is FALSE, then R will not evaluate condition 2. && only works for single logical values - not vectors.
slwr <- function() {
for(i in 1:10000) {
x <- rnorm(1);
if(x > .4 & x < .6) {y <- x}
}
}
fstr <- function() {
for(i in 1:10000) {
x <- rnorm(1);
if(x > .4 && x < .6) {y <- x}
}
}
microbenchmark(slwr, fstr, times = 10)
## Warning in microbenchmark(slwr, fstr, times = 10): Could not measure a positive
## execution time for 3 evaluations.
## Unit: nanoseconds
## expr min lq mean median uq max neval cld
## slwr 0 0 60 0 0 500 10 a
## fstr 0 0 300 0 0 3000 10 a3.2 Defensive Coding
These notes are from the DataCamp course Writing Efficient R Code.↩︎