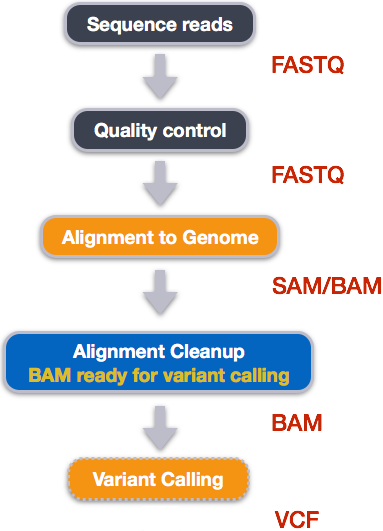
5 Variant Calling Workflow
Sys.setenv(DCWORKSHOP='~/dc_workshop')Now we write a bash code chunk:
echo $DCWORKSHOP
pwd
cd $DCWORKSHOP
pwd## ~/dc_workshop
## /Users/ggiaever/Documents/RProjects/genomics/wrangling-genomics
## zsh:cd:3: no such file or directory: ~/dc_workshop
## /Users/ggiaever/Documents/RProjects/genomics/wrangling-genomicsWe mentioned before that we are working with files from a long-term evolution study of an E. coli population (designated Ara-3). Now that we have looked at our data to make sure that it is high quality, and removed low-quality base calls, we can perform variant calling to see how the population changed over time. We care how this population changed relative to the original population, E. coli strain REL606. Therefore, we will align each of our samples to the E. coli REL606 reference genome, and see what differences exist in our reads versus the genome.
5.1 Alignment to a reference genome
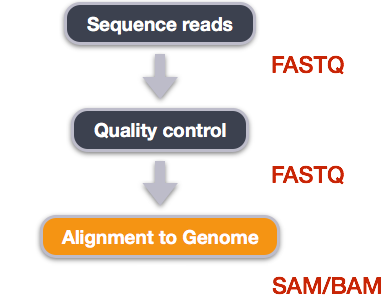
We perform read alignment or mapping to determine where in the genome our reads originated from. There are a number of tools to choose from and, while there is no gold standard, there are some tools that are better suited for particular NGS analyses. We will be using the Burrows Wheeler Aligner (BWA), which is a software package for mapping low-divergent sequences against a large reference genome.
The alignment process consists of two steps:
- Indexing the reference genome
- Aligning the reads to the reference genome
5.2 Setting up
First we download the reference genome for E. coli REL606. Although we could copy or move the file with cp or mv, most genomics workflows begin with a download step, so we will practice that here.
cd ~/dc_workshop
mkdir -p data/ref_genome
curl -L -o data/ref_genome/ecoli_rel606.fasta.gz ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCA/000/017/985/GCA_000017985.1_ASM1798v1/GCA_000017985.1_ASM1798v1_genomic.fna.gz
gunzip -f data/ref_genome/ecoli_rel606.fasta.gz## % Total % Received % Xferd Average Speed Time Time Time Current
## Dload Upload Total Spent Left Speed
##
0 0 0 0 0 0 0 0 --:--:-- --:--:-- --:--:-- 0
0 0 0 0 0 0 0 0 --:--:-- --:--:-- --:--:-- 0
0 0 0 0 0 0 0 0 --:--:-- 0:00:01 --:--:-- 0
11 1343k 11 156k 0 0 72021 0 0:00:19 0:00:02 0:00:17 72172
40 1343k 40 540k 0 0 167k 0 0:00:08 0:00:03 0:00:05 167k
98 1343k 98 1319k 0 0 311k 0 0:00:04 0:00:04 --:--:-- 312k
100 1343k 100 1343k 0 0 310k 0 0:00:04 0:00:04 --:--:-- 330k5.3 Exercise
We saved this file as data/ref_genome/ecoli_rel606.fasta.gz and then decompressed it.
What is the real name of the genome?
5.4 Solution
We will also download a set of trimmed FASTQ files to work with. These are small subsets of our real trimmed data, and will enable us to run our variant calling workflow quite quickly.
I couldn’t get this to work – just do it manually –
cd ~/dc_workshop
curl L -o sub.tar.gz https://ndownloader.figshare.com/files/14418248
mkdir -p /dc_workshop/data/trimmed_fastq_small
cd ~/dc_workshop/data/trimmed_fastq_small
tar -xvf sub.tar.gz
mv -v sub/* ../trimmed_fastq_smallYou will also need to create directories for the results that will be generated as part of this workflow. We can do this in a single
line of code, because mkdir can accept multiple new directory
names as input.
cd ~/dc_workshop/results
mkdir -p sam bam bcf vcf5.4.1 Index the reference genome
Our first step is to index the reference genome for use by BWA. Indexing allows the aligner to quickly find potential alignment sites for query sequences in a genome, which saves time during alignment. Indexing the reference only has to be run once. The only reason you would want to create a new index is if you are working with a different reference genome or you are using a different tool for alignment.
cd ~/dc_workshop
source $HOME/miniconda3/bin/activate
conda activate variant
bwa index data/ref_genome/ecoli_rel606.fasta## [bwa_index] Pack FASTA... 0.03 sec
## [bwa_index] Construct BWT for the packed sequence...
## [bwa_index] 0.64 seconds elapse.
## [bwa_index] Update BWT... 0.02 sec
## [bwa_index] Pack forward-only FASTA... 0.02 sec
## [bwa_index] Construct SA from BWT and Occ... 0.20 sec
## [main] Version: 0.7.17-r1188
## [main] CMD: bwa index data/ref_genome/ecoli_rel606.fasta
## [main] Real time: 0.903 sec; CPU: 0.925 secWhile the index is created, you will see output that looks something like this:
[bwa_index] Pack FASTA… 0.04 sec [bwa_index] Construct BWT for the packed sequence… [bwa_index] 1.05 seconds elapse. [bwa_index] Update BWT… 0.03 sec [bwa_index] Pack forward-only FASTA… 0.02 sec [bwa_index] Construct SA from BWT and Occ… 0.57 sec [main] Version: 0.7.17-r1188 [main] CMD: bwa index data/ref_genome/ecoli_rel606.fasta [main] Real time: 1.765 sec; CPU: 1.715 sec
5.4.2 Align reads to reference genome
The alignment process consists of choosing an appropriate reference genome to map our reads against and then deciding on an aligner. We will use the BWA-MEM algorithm, which is the latest and is generally recommended for high-quality queries as it is faster and more accurate.
An example of what a bwa command looks like is below. This command will not run, as we do not have the files ref_genome.fa, input_file_R1.fastq, or input_file_R2.fastq.
cd ~/dc_workshop
source $HOME/miniconda3/bin/activate
conda activate variant
bwa mem ref_genome.fasta input_file_R1.fastq input_file_R2.fastq > output.samHave a look at the bwa options page. While we are running bwa with the default parameters here, your use case might require a change of parameters. NOTE: Always read the manual page for any tool before using and make sure the options you use are appropriate for your data.
We are going to start by aligning the reads from just one of the
samples in our dataset (SRR2584866). Later, we will be
iterating this whole process on all of our sample files.
cd ~/dc_workshop
source $HOME/miniconda3/bin/activate
conda activate variant
bwa mem data/ref_genome/ecoli_rel606.fasta data/trimmed_fastq_small/SRR2584866_1.trim.sub.fastq data/trimmed_fastq_small/SRR2584866_2.trim.sub.fastq > results/sam/SRR2584866.aligned.sam## [M::bwa_idx_load_from_disk] read 0 ALT contigs
## [M::process] read 77446 sequences (10000033 bp)...
## [M::process] read 77296 sequences (10000182 bp)...
## [M::mem_pestat] # candidate unique pairs for (FF, FR, RF, RR): (48, 36728, 21, 61)
## [M::mem_pestat] analyzing insert size distribution for orientation FF...
## [M::mem_pestat] (25, 50, 75) percentile: (420, 660, 1774)
## [M::mem_pestat] low and high boundaries for computing mean and std.dev: (1, 4482)
## [M::mem_pestat] mean and std.dev: (784.68, 700.87)
## [M::mem_pestat] low and high boundaries for proper pairs: (1, 5836)
## [M::mem_pestat] analyzing insert size distribution for orientation FR...
## [M::mem_pestat] (25, 50, 75) percentile: (221, 361, 576)
## [M::mem_pestat] low and high boundaries for computing mean and std.dev: (1, 1286)
## [M::mem_pestat] mean and std.dev: (412.89, 227.06)
## [M::mem_pestat] low and high boundaries for proper pairs: (1, 1641)
## [M::mem_pestat] analyzing insert size distribution for orientation RF...
## [M::mem_pestat] (25, 50, 75) percentile: (560, 2011, 2594)
## [M::mem_pestat] low and high boundaries for computing mean and std.dev: (1, 6662)
## [M::mem_pestat] mean and std.dev: (1580.30, 978.54)
## [M::mem_pestat] low and high boundaries for proper pairs: (1, 8696)
## [M::mem_pestat] analyzing insert size distribution for orientation RR...
## [M::mem_pestat] (25, 50, 75) percentile: (320, 549, 942)
## [M::mem_pestat] low and high boundaries for computing mean and std.dev: (1, 2186)
## [M::mem_pestat] mean and std.dev: (581.31, 431.43)
## [M::mem_pestat] low and high boundaries for proper pairs: (1, 2808)
## [M::mem_pestat] skip orientation FF
## [M::mem_pestat] skip orientation RF
## [M::mem_pestat] skip orientation RR
## [M::mem_process_seqs] Processed 77446 reads in 2.016 CPU sec, 1.980 real sec
## [M::process] read 75458 sequences (10000048 bp)...
## [M::mem_pestat] # candidate unique pairs for (FF, FR, RF, RR): (65, 36742, 10, 60)
## [M::mem_pestat] analyzing insert size distribution for orientation FF...
## [M::mem_pestat] (25, 50, 75) percentile: (172, 452, 1166)
## [M::mem_pestat] low and high boundaries for computing mean and std.dev: (1, 3154)
## [M::mem_pestat] mean and std.dev: (645.66, 709.63)
## [M::mem_pestat] low and high boundaries for proper pairs: (1, 4148)
## [M::mem_pestat] analyzing insert size distribution for orientation FR...
## [M::mem_pestat] (25, 50, 75) percentile: (224, 368, 582)
## [M::mem_pestat] low and high boundaries for computing mean and std.dev: (1, 1298)
## [M::mem_pestat] mean and std.dev: (417.93, 229.24)
## [M::mem_pestat] low and high boundaries for proper pairs: (1, 1656)
## [M::mem_pestat] analyzing insert size distribution for orientation RF...
## [M::mem_pestat] (25, 50, 75) percentile: (2317, 2353, 2422)
## [M::mem_pestat] low and high boundaries for computing mean and std.dev: (2107, 2632)
## [M::mem_pestat] mean and std.dev: (2319.12, 75.27)
## [M::mem_pestat] low and high boundaries for proper pairs: (2002, 2737)
## [M::mem_pestat] analyzing insert size distribution for orientation RR...
## [M::mem_pestat] (25, 50, 75) percentile: (244, 513, 1366)
## [M::mem_pestat] low and high boundaries for computing mean and std.dev: (1, 3610)
## [M::mem_pestat] mean and std.dev: (781.13, 858.61)
## [M::mem_pestat] low and high boundaries for proper pairs: (1, 4732)
## [M::mem_pestat] skip orientation FF
## [M::mem_pestat] skip orientation RF
## [M::mem_pestat] skip orientation RR
## [M::mem_process_seqs] Processed 77296 reads in 2.029 CPU sec, 1.958 real sec
## [M::process] read 74054 sequences (10000155 bp)...
## [M::mem_pestat] # candidate unique pairs for (FF, FR, RF, RR): (58, 35818, 13, 79)
## [M::mem_pestat] analyzing insert size distribution for orientation FF...
## [M::mem_pestat] (25, 50, 75) percentile: (288, 553, 1479)
## [M::mem_pestat] low and high boundaries for computing mean and std.dev: (1, 3861)
## [M::mem_pestat] mean and std.dev: (822.66, 856.80)
## [M::mem_pestat] low and high boundaries for proper pairs: (1, 5052)
## [M::mem_pestat] analyzing insert size distribution for orientation FR...
## [M::mem_pestat] (25, 50, 75) percentile: (224, 361, 576)
## [M::mem_pestat] low and high boundaries for computing mean and std.dev: (1, 1280)
## [M::mem_pestat] mean and std.dev: (414.99, 227.46)
## [M::mem_pestat] low and high boundaries for proper pairs: (1, 1632)
## [M::mem_pestat] analyzing insert size distribution for orientation RF...
## [M::mem_pestat] (25, 50, 75) percentile: (1176, 2230, 2643)
## [M::mem_pestat] low and high boundaries for computing mean and std.dev: (1, 5577)
## [M::mem_pestat] mean and std.dev: (1799.00, 921.05)
## [M::mem_pestat] low and high boundaries for proper pairs: (1, 7044)
## [M::mem_pestat] analyzing insert size distribution for orientation RR...
## [M::mem_pestat] (25, 50, 75) percentile: (365, 601, 1880)
## [M::mem_pestat] low and high boundaries for computing mean and std.dev: (1, 4910)
## [M::mem_pestat] mean and std.dev: (1144.51, 1264.10)
## [M::mem_pestat] low and high boundaries for proper pairs: (1, 6425)
## [M::mem_pestat] skip orientation FF
## [M::mem_pestat] skip orientation RF
## [M::mem_pestat] skip orientation RR
## [M::mem_process_seqs] Processed 75458 reads in 2.055 CPU sec, 1.986 real sec
## [M::process] read 45746 sequences (6219545 bp)...
## [M::mem_pestat] # candidate unique pairs for (FF, FR, RF, RR): (41, 35149, 14, 68)
## [M::mem_pestat] analyzing insert size distribution for orientation FF...
## [M::mem_pestat] (25, 50, 75) percentile: (321, 488, 1274)
## [M::mem_pestat] low and high boundaries for computing mean and std.dev: (1, 3180)
## [M::mem_pestat] mean and std.dev: (607.09, 527.87)
## [M::mem_pestat] low and high boundaries for proper pairs: (1, 4133)
## [M::mem_pestat] analyzing insert size distribution for orientation FR...
## [M::mem_pestat] (25, 50, 75) percentile: (220, 353, 566)
## [M::mem_pestat] low and high boundaries for computing mean and std.dev: (1, 1258)
## [M::mem_pestat] mean and std.dev: (407.78, 223.19)
## [M::mem_pestat] low and high boundaries for proper pairs: (1, 1604)
## [M::mem_pestat] analyzing insert size distribution for orientation RF...
## [M::mem_pestat] (25, 50, 75) percentile: (652, 2237, 2446)
## [M::mem_pestat] low and high boundaries for computing mean and std.dev: (1, 6034)
## [M::mem_pestat] mean and std.dev: (1704.23, 991.13)
## [M::mem_pestat] low and high boundaries for proper pairs: (1, 7828)
## [M::mem_pestat] analyzing insert size distribution for orientation RR...
## [M::mem_pestat] (25, 50, 75) percentile: (231, 409, 1018)
## [M::mem_pestat] low and high boundaries for computing mean and std.dev: (1, 2592)
## [M::mem_pestat] mean and std.dev: (513.68, 468.59)
## [M::mem_pestat] low and high boundaries for proper pairs: (1, 3379)
## [M::mem_pestat] skip orientation FF
## [M::mem_pestat] skip orientation RF
## [M::mem_pestat] skip orientation RR
## [M::mem_process_seqs] Processed 74054 reads in 2.035 CPU sec, 1.975 real sec
## [M::mem_pestat] # candidate unique pairs for (FF, FR, RF, RR): (40, 21701, 9, 32)
## [M::mem_pestat] analyzing insert size distribution for orientation FF...
## [M::mem_pestat] (25, 50, 75) percentile: (235, 720, 1382)
## [M::mem_pestat] low and high boundaries for computing mean and std.dev: (1, 3676)
## [M::mem_pestat] mean and std.dev: (770.19, 676.46)
## [M::mem_pestat] low and high boundaries for proper pairs: (1, 4823)
## [M::mem_pestat] analyzing insert size distribution for orientation FR...
## [M::mem_pestat] (25, 50, 75) percentile: (219, 350, 560)
## [M::mem_pestat] low and high boundaries for computing mean and std.dev: (1, 1242)
## [M::mem_pestat] mean and std.dev: (404.41, 221.74)
## [M::mem_pestat] low and high boundaries for proper pairs: (1, 1583)
## [M::mem_pestat] skip orientation RF as there are not enough pairs
## [M::mem_pestat] analyzing insert size distribution for orientation RR...
## [M::mem_pestat] (25, 50, 75) percentile: (325, 557, 721)
## [M::mem_pestat] low and high boundaries for computing mean and std.dev: (1, 1513)
## [M::mem_pestat] mean and std.dev: (501.66, 309.62)
## [M::mem_pestat] low and high boundaries for proper pairs: (1, 1909)
## [M::mem_pestat] skip orientation FF
## [M::mem_pestat] skip orientation RR
## [M::mem_process_seqs] Processed 45746 reads in 1.311 CPU sec, 1.271 real sec
## [main] Version: 0.7.17-r1188
## [main] CMD: bwa mem data/ref_genome/ecoli_rel606.fasta data/trimmed_fastq_small/SRR2584866_1.trim.sub.fastq data/trimmed_fastq_small/SRR2584866_2.trim.sub.fastq
## [main] Real time: 9.243 sec; CPU: 9.539 secYou will see output that starts like this:
[M::bwa_idx_load_from_disk] read 0 ALT contigs [M::process] read 77446 sequences (10000033 bp)… [M::process] read 77296 sequences (10000182 bp)… [M::mem_pestat] # candidate unique pairs for (FF, FR, RF, RR): (48, 36728, 21, 61) [M::mem_pestat] analyzing insert size distribution for orientation FF… [M::mem_pestat] (25, 50, 75) percentile: (420, 660, 1774) [M::mem_pestat] low and high boundaries for computing mean and std.dev: (1, 4482) [M::mem_pestat] mean and std.dev: (784.68, 700.87) [M::mem_pestat] low and high boundaries for proper pairs: (1, 5836) [M::mem_pestat] analyzing insert size distribution for orientation
5.4.2.1 SAM/BAM format
The SAM file, is a tab-delimited text file that contains information for each individual read and its alignment to the genome. While we do not have time to go into detail about the features of the SAM format, the paper by Heng Li et al. provides a lot more detail on the specification.
The compressed binary version of SAM is called a BAM file. We use this version to reduce size and to allow for indexing, which enables efficient random access of the data contained within the file.
The file begins with a header, which is optional. The header is used to describe the source of data, reference sequence, method of alignment, etc., this will change depending on the aligner being used. Following the header is the alignment section. Each line that follows corresponds to alignment information for a single read. Each alignment line has 11 mandatory fields for essential mapping information and a variable number of other fields for aligner specific information. An example entry from a SAM file is displayed below with the different fields highlighted.
Additionally tags (or attribute) can be added to each of of the lines. These tags give some additional information on the alignment. The number and type of tags varies between different alignment tools and the settings within these tools. The ‘CIGAR’ (Compact Idiosyncratic Gapped Alignment Report) string is how the SAM/BAM format represents spliced alignments. Understanding the CIGAR string will help you understand how your query sequence aligns to the reference genome. For example, the position stored is the left most coordinate of the alignment. To get to the right coordinate, you have to parse the CIGAR string.
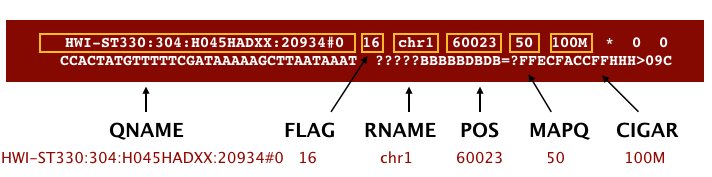
The sam/bam mandatory tags: 1-6
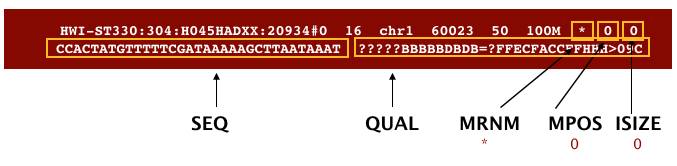
The sam/bam mandatory tags: 7-11
We will convert the SAM file to BAM format using the samtools program with the view command and tell this command that the input is in SAM format (-S) and to output BAM format (-b):
cd ~/dc_workshop
source $HOME/miniconda3/bin/activate
conda activate variant
samtools view -S -b results/sam/SRR2584866.aligned.sam > results/bam/SRR2584866.aligned.bam11 mandatory fields
library(Rsamtools)## Loading required package: GenomeInfoDb## Warning: package 'GenomeInfoDb' was built under R version 4.2.2## Loading required package: BiocGenerics##
## Attaching package: 'BiocGenerics'## The following objects are masked from 'package:stats':
##
## IQR, mad, sd, var, xtabs## The following objects are masked from 'package:base':
##
## anyDuplicated, aperm, append, as.data.frame, basename, cbind,
## colnames, dirname, do.call, duplicated, eval, evalq, Filter, Find,
## get, grep, grepl, intersect, is.unsorted, lapply, Map, mapply,
## match, mget, order, paste, pmax, pmax.int, pmin, pmin.int,
## Position, rank, rbind, Reduce, rownames, sapply, setdiff, sort,
## table, tapply, union, unique, unsplit, which.max, which.min## Loading required package: S4Vectors## Loading required package: stats4##
## Attaching package: 'S4Vectors'## The following objects are masked from 'package:base':
##
## expand.grid, I, unname## Loading required package: IRanges## Loading required package: GenomicRanges## Loading required package: Biostrings## Loading required package: XVector##
## Attaching package: 'Biostrings'## The following object is masked from 'package:base':
##
## strsplitf1 = "/Users/ggiaever/dc_workshop/results/bam/SRR2584866.aligned.bam"
reads = scanBam(BamFile("/Users/ggiaever/dc_workshop/results/bam/SRR2584866.aligned.bam",yieldSize = 20))
n = reads[[1]]
names(n)## [1] "qname" "flag" "rname" "strand" "pos" "qwidth" "mapq" "cigar"
## [9] "mrnm" "mpos" "isize" "seq" "qual"nchar(n$seq)## [1] 26 150 26 150 27 108 25 50 67 68 133 100 129 90 50 150 150 150 150
## [20] 46n$seq## DNAStringSet object of length 20:
## width seq
## [1] 26 GCTTCCCATGCCAATTAATACATGTG
## [2] 150 TCCTCAATCTTAAGCGGATCAATGAGCTGGTAC...CGCCTTTCACCCCTGCCATACCTTTTAATAATC
## [3] 26 GCTGATATTCTTGCAGCAGTACCGGC
## [4] 150 ATAGCCGCCGTTTCTTTTTGTGCACCGGCGGCA...GTTGATGCCTGATACACCTGTTCCATTAAAGGT
## [5] 27 GTTATCAGCGAGTTCCTGATTAAACGA
## ... ... ...
## [16] 150 ATGTATAACAATAAGTTGCAAAGCACGTTGCAT...GCTGGCGATTCGGGCTAGTGTCCACTCATGGGC
## [17] 150 GTTTTATGTTCAGATAATGCCCGATGACTTTGT...CAATTCGCTGCGTATATCGCTTGCTGATTACGT
## [18] 150 CGTCTTCCGGAGCCTGTCATACGCGTAAAACAG...GCGCGAGGTTACCGACTGCGGCCTGAGTTTTTT
## [19] 150 GCAAAACCCTTTTTTGAACGTTATGGTTTTCAG...CGTTGAACGTTATATCGATATGTTATTTGTTGA
## [20] 46 GTTTTATTACCTGCGTTGAACGTTATATCGATATGTTATTTGTTGAcomplement(n$seq)## DNAStringSet object of length 20:
## width seq
## [1] 26 CGAAGGGTACGGTTAATTATGTACAC
## [2] 150 AGGAGTTAGAATTCGCCTAGTTACTCGACCATG...GCGGAAAGTGGGGACGGTATGGAAAATTATTAG
## [3] 26 CGACTATAAGAACGTCGTCATGGCCG
## [4] 150 TATCGGCGGCAAAGAAAAACACGTGGCCGCCGT...CAACTACGGACTATGTGGACAAGGTAATTTCCA
## [5] 27 CAATAGTCGCTCAAGGACTAATTTGCT
## ... ... ...
## [16] 150 TACATATTGTTATTCAACGTTTCGTGCAACGTA...CGACCGCTAAGCCCGATCACAGGTGAGTACCCG
## [17] 150 CAAAATACAAGTCTATTACGGGCTACTGAAACA...GTTAAGCGACGCATATAGCGAACGACTAATGCA
## [18] 150 GCAGAAGGCCTCGGACAGTATGCGCATTTTGTC...CGCGCTCCAATGGCTGACGCCGGACTCAAAAAA
## [19] 150 CGTTTTGGGAAAAAACTTGCAATACCAAAAGTC...GCAACTTGCAATATAGCTATACAATAAACAACT
## [20] 46 CAAAATAATGGACGCAACTTGCAATATAGCTATACAATAAACAACTn$strand## [1] - + + - + - + - + - + - + - - + + - + +
## Levels: + - *n$pos## [1] 3585268 3584643 2861836 2862333 1550866 1550866 337306 337306 2813980
## [10] 2814304 2934043 2934475 504911 505227 3594815 3594687 589581 589867
## [19] 257614 257477n$flag## [1] 83 163 99 147 99 147 99 147 99 147 99 147 99 147 83
## [16] 163 99 147 99 2147n$qname## [1] "SRR2584866.7" "SRR2584866.7" "SRR2584866.8" "SRR2584866.8"
## [5] "SRR2584866.14" "SRR2584866.14" "SRR2584866.15" "SRR2584866.15"
## [9] "SRR2584866.21" "SRR2584866.21" "SRR2584866.23" "SRR2584866.23"
## [13] "SRR2584866.24" "SRR2584866.24" "SRR2584866.26" "SRR2584866.26"
## [17] "SRR2584866.27" "SRR2584866.27" "SRR2584866.29" "SRR2584866.29"n$qwidth## [1] 26 150 26 150 27 108 25 50 67 68 133 100 129 90 50 150 150 150 150
## [20] 46n$qual## PhredQuality object of length 20:
## width seq
## [1] 26 IJJJJJJJJJIJJHHHHHFEFFFCC@
## [2] 150 C@CFFFFFGHGGGGGJJJIJJJJIJJJIJIHII...DDDDBDDCDDDDDBDDBDDDDDDDDDDDDEDED
## [3] 26 @@@DBDDDHHFFHIFFGBH<FHCGDG
## [4] 150 C@DDD@DDDDDDBBBDDADDDDDBDDDDDDDCD...IFJJJJJGIIIGHEHEIIJJHHGHHFFFFFCC@
## [5] 27 CCCFFFFFHHHHHJJJJJIJJJJJJJJ
## ... ... ...
## [16] 150 CCCFFFFFHHGHHJJHIJIJJJIIJJJHIIIHI...D>?B@B@3>ACDD@<9BBCCCD>>>CA?CCD?B
## [17] 150 C@CFFFFFHHHHHJJJJJJJJIJIIJJJJJJII...<@ACDC@@BBDBBDBDCC?BDBDBCCACDCCDD
## [18] 150 BCAB>DDD@A8B@DCC@A<@<@@<DCCDCBDDD...JGJJJIJJJHIJJJJJJJJJHHHHHFFFFFCCC
## [19] 150 @CCFFFFFHHHHHJJIJJJIHIGIIJHIJJJII...DDDDDDCCABCDDEEEDDDDDDEDEEDEEEDDC
## [20] 46 BDDEDDDDEDCDDDDDDDDCCABCDDEEEDDDDDDEDEEDEEEDDCn$cigar## [1] "26M" "150M" "26M" "150M" "27M" "108M" "25M"
## [8] "50M" "67M" "68M" "133M" "100M" "129M" "90M"
## [15] "50M" "150M" "150M" "150M" "104M46S" "104H46M"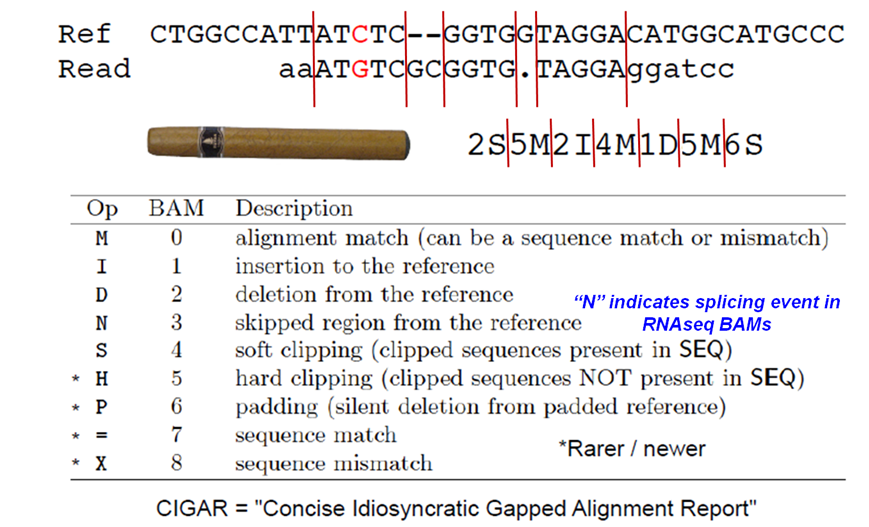 ### Sort BAM file by coordinates
### Sort BAM file by coordinates
Next we sort the BAM file using the sort command from samtools. -o tells the command where to write the output.
cd ~/dc_workshop
source $HOME/miniconda3/bin/activate
conda activate variant
samtools sort -o results/bam/SRR2584866.aligned.sorted.bam results/bam/SRR2584866.aligned.bam Our files are pretty small, so we will not see this output. If you run the workflow with larger files, you will see something like this:
[bam_sort_core] merging from 2 files…
SAM/BAM files can be sorted in multiple ways, e.g. by location of alignment on the chromosome, by read name, etc. It is important to be aware that different alignment tools will output differently sorted SAM/BAM, and different downstream tools require differently sorted alignment files as input.
You can use samtools to learn more about this bam file as well.
cd ~/dc_workshop
source $HOME/miniconda3/bin/activate
conda activate variant
samtools flagstat results/bam/SRR2584866.aligned.sorted.bam## 351169 + 0 in total (QC-passed reads + QC-failed reads)
## 350000 + 0 primary
## 0 + 0 secondary
## 1169 + 0 supplementary
## 0 + 0 duplicates
## 0 + 0 primary duplicates
## 351103 + 0 mapped (99.98% : N/A)
## 349934 + 0 primary mapped (99.98% : N/A)
## 350000 + 0 paired in sequencing
## 175000 + 0 read1
## 175000 + 0 read2
## 346688 + 0 properly paired (99.05% : N/A)
## 349876 + 0 with itself and mate mapped
## 58 + 0 singletons (0.02% : N/A)
## 0 + 0 with mate mapped to a different chr
## 0 + 0 with mate mapped to a different chr (mapQ>=5)This will give you the following statistics about your sorted bam file:
351169 + 0 in total (QC-passed reads + QC-failed reads) 0 + 0 secondary 1169 + 0 supplementary 0 + 0 duplicates 351103 + 0 mapped (99.98% : N/A) 350000 + 0 paired in sequencing 175000 + 0 read1 175000 + 0 read2 346688 + 0 properly paired (99.05% : N/A) 349876 + 0 with itself and mate mapped 58 + 0 singletons (0.02% : N/A) 0 + 0 with mate mapped to a different chr 0 + 0 with mate mapped to a different chr (mapQ=5)
5.5 Variant calling
A variant call is a conclusion that there is a nucleotide difference vs. some reference at a given position in an individual genome
or transcriptome, often referred to as a Single Nucleotide Variant (SNV). The call is usually accompanied by an estimate of
variant frequency and some measure of confidence. Similar to other steps in this workflow, there are a number of tools available for
variant calling. In this workshop we will be using bcftools, but there are a few things we need to do before actually calling the
variants.
5.5.1 Step 1: Calculate the read coverage of positions in the genome
Do the first pass on variant calling by counting read coverage with
bcftools. We will
use the command mpileup. The flag -O b tells bcftools to generate a
bcf format output file, -o specifies where to write the output file, and -f flags the path to the reference genome:
cd ~/dc_workshop
source $HOME/miniconda3/bin/activate
conda activate variant
bcftools mpileup -O b -o results/bcf/SRR2584866_raw.bcf \
-f data/ref_genome/ecoli_rel606.fasta results/bam/SRR2584866.aligned.sorted.bam## [mpileup] 1 samples in 1 input files
## [mpileup] maximum number of reads per input file set to -d 250We have now generated a file with coverage information for every base.
5.5.2 Step 2: Detect the single nucleotide variants (SNVs)
Identify SNVs using bcftools call. We have to specify ploidy with the flag --ploidy, which is one for the haploid E. coli. -m allows for multiallelic and rare-variant calling, -v tells the program to output variant sites only (not every site in the genome), and -o specifies where to write the output file:
cd ~/dc_workshop
source $HOME/miniconda3/bin/activate
conda activate variant
bcftools call --ploidy 1 -m -v -o results/vcf/SRR2584866_variants.vcf results/bcf/SRR2584866_raw.bcf 5.5.3 Step 3: Filter and report the SNV variants in variant calling format (VCF)
To filter out SNPs that are low quality or covered by low depth, we can use the vcfutils.pl varFilter that comes with samtools. There are a lot of options you can use, like read depth, mapping quality but for now we will just go with the defaults, For depth you can use the -d option and for quality you can use the -Q option. Try playing around with that.
Filter the SNVs for the final output in VCF format, using vcfutils.pl:
cd ~/dc_workshop
source $HOME/miniconda3/bin/activate
conda activate variant
vcfutils.pl varFilter results/vcf/SRR2584866_variants.vcf > results/vcf/SRR2584866_final_variants.vcf
cd ~/dc_workshop
less -S results/vcf/SRR2584866_final_variants.vcfYou will see the header (which describes the format), the time and date the file was created, the version of bcftools that was used, the command line parameters used, and some additional information:
fileformat=VCFv4.2 FILTER=<ID=PASS,Description=“All filters passed” bcftoolsVersion=1.8+htslib-1.8 bcftoolsCommand=mpileup -O b -o results/bcf/SRR2584866_raw.bcf -f data/ref_genome/ecoli_rel606.fasta results/bam/SRR2584866.aligned.sorted.bam reference=file://data/ref_genome/ecoli_rel606.fasta contig=<ID=CP000819.1,length=4629812 ALT=<ID=*,Description=“Represents allele(s) other than observed.” INFO=<ID=INDEL,Number=0,Type=Flag,Description=“Indicates that the variant is an INDEL.” INFO=<ID=IDV,Number=1,Type=Integer,Description=“Maximum number of reads supporting an indel” INFO=<ID=IMF,Number=1,Type=Float,Description=“Maximum fraction of reads supporting an indel” INFO=<ID=DP,Number=1,Type=Integer,Description=“Raw read depth” INFO=<ID=VDB,Number=1,Type=Float,Description=“Variant Distance Bias for filtering splice-site artefacts in RNA-seq data (bigger is better)”,Version= INFO=<ID=RPB,Number=1,Type=Float,Description=“Mann-Whitney U test of Read Position Bias (bigger is better)” INFO=<ID=MQB,Number=1,Type=Float,Description=“Mann-Whitney U test of Mapping Quality Bias (bigger is better)” INFO=<ID=BQB,Number=1,Type=Float,Description=“Mann-Whitney U test of Base Quality Bias (bigger is better)” INFO=<ID=MQSB,Number=1,Type=Float,Description=“Mann-Whitney U test of Mapping Quality vs Strand Bias (bigger is better)” INFO=<ID=SGB,Number=1,Type=Float,Description=“Segregation based metric.” INFO=<ID=MQ0F,Number=1,Type=Float,Description=“Fraction of MQ0 reads (smaller is better)” FORMAT=<ID=PL,Number=G,Type=Integer,Description=“List of Phred-scaled genotype likelihoods” FORMAT=<ID=GT,Number=1,Type=String,Description=“Genotype” INFO=<ID=ICB,Number=1,Type=Float,Description=“Inbreeding Coefficient Binomial test (bigger is better)” INFO=<ID=HOB,Number=1,Type=Float,Description=“Bias in the number of HOMs number (smaller is better)” INFO=<ID=AC,Number=A,Type=Integer,Description=“Allele count in genotypes for each ALT allele, in the same order as listed” INFO=<ID=AN,Number=1,Type=Integer,Description=“Total number of alleles in called genotypes” INFO=<ID=DP4,Number=4,Type=Integer,Description=“Number of high-quality ref-forward , ref-reverse, alt-forward and alt-reverse bases” INFO=<ID=MQ,Number=1,Type=Integer,Description=“Average mapping quality” bcftools_callVersion=1.8+htslib-1.8 bcftools_callCommand=call –ploidy 1 -m -v -o results/bcf/SRR2584866_variants.vcf results/bcf/SRR2584866_raw.bcf; Date=Tue Oct 9 18:48:10 2018
Followed by information on each of the variations observed:
CHROM POS ID REF ALT QUAL FILTER INFO FORMAT results/bam/SRR2584866.aligned.sorted.bam CP000819.1 1521 . C T 207 . DP=9;VDB=0.993024;SGB=-0.662043;MQSB=0.974597;MQ0F=0;AC=1;AN=1;DP4=0,0,4,5;MQ=60 CP000819.1 1612 . A G 225 . DP=13;VDB=0.52194;SGB=-0.676189;MQSB=0.950952;MQ0F=0;AC=1;AN=1;DP4=0,0,6,5;MQ=60 CP000819.1 9092 . A G 225 . DP=14;VDB=0.717543;SGB=-0.670168;MQSB=0.916482;MQ0F=0;AC=1;AN=1;DP4=0,0,7,3;MQ=60 CP000819.1 9972 . T G 214 . DP=10;VDB=0.022095;SGB=-0.670168;MQSB=1;MQ0F=0;AC=1;AN=1;DP4=0,0,2,8;MQ=60 GT:PL CP000819.1 10563 . G A 225 . DP=11;VDB=0.958658;SGB=-0.670168;MQSB=0.952347;MQ0F=0;AC=1;AN=1;DP4=0,0,5,5;MQ=60 CP000819.1 22257 . C T 127 . DP=5;VDB=0.0765947;SGB=-0.590765;MQSB=1;MQ0F=0;AC=1;AN=1;DP4=0,0,2,3;MQ=60 GT:PL CP000819.1 38971 . A G 225 . DP=14;VDB=0.872139;SGB=-0.680642;MQSB=1;MQ0F=0;AC=1;AN=1;DP4=0,0,4,8;MQ=60 GT:PL CP000819.1 42306 . A G 225 . DP=15;VDB=0.969686;SGB=-0.686358;MQSB=1;MQ0F=0;AC=1;AN=1;DP4=0,0,5,9;MQ=60 GT:PL CP000819.1 45277 . A G 225 . DP=15;VDB=0.470998;SGB=-0.680642;MQSB=0.95494;MQ0F=0;AC=1;AN=1;DP4=0,0,7,5;MQ=60 CP000819.1 56613 . C G 183 . DP=12;VDB=0.879703;SGB=-0.676189;MQSB=1;MQ0F=0;AC=1;AN=1;DP4=0,0,8,3;MQ=60 GT:PL CP000819.1 62118 . A G 225 . DP=19;VDB=0.414981;SGB=-0.691153;MQSB=0.906029;MQ0F=0;AC=1;AN=1;DP4=0,0,8,10;MQ=59 CP000819.1 64042 . G A 225 . DP=18;VDB=0.451328;SGB=-0.689466;MQSB=1;MQ0F=0;AC=1;AN=1;DP4=0,0,7,9;MQ=60 GT:PL
This is a lot of information, so let’s take some time to make sure we understand our output.
The first few columns represent the information we have about a predicted variation.
| column | info |
|---|---|
| CHROM | contig location where the variation occurs |
| POS | position within the contig where the variation occurs |
| ID | a . until we add annotation information |
| REF | reference genotype (forward strand) |
| ALT | sample genotype (forward strand) |
| QUAL | Phred-scaled probability that the observed variant exists at this site (higher is better) |
| FILTER | a . if no quality filters have been applied, PASS if a filter is passed, or the name of the filters this variant failed |
In an ideal world, the information in the QUAL column would be all we needed to filter out bad variant calls.
However, in reality we need to filter on multiple other metrics.
The last two columns contain the genotypes and can be tricky to decode.
| column | info |
|---|---|
| FORMAT | lists in order the metrics presented in the final column |
| results | lists the values associated with those metrics in order |
For our file, the metrics presented are GT:PL:GQ.
| metric | definition |
|---|---|
| AD, DP | the depth per allele by sample and coverage |
| GT | the genotype for the sample at this loci. For a diploid organism, the GT field indicates the two alleles carried by the sample, encoded by a 0 for the REF allele, 1 for the first ALT allele, 2 for the second ALT allele, etc. A 0/0 means homozygous reference, 0/1 is heterozygous, and 1/1 is homozygous for the alternate allele. |
| PL | the likelihoods of the given genotypes |
| GQ | the Phred-scaled confidence for the genotype |
The Broad Institute’s VCF guide is an excellent place to learn more about the VCF file format.
5.6 Exercise
Use the grep and wc commands you have learned to assess how many variants are in the vcf file.
5.8 Assess the alignment (visualization) - optional step
It is often instructive to look at your data in a genome browser. Visualization will allow you to get a “feel” for the data, as well as detecting abnormalities and problems. Also, exploring the data in such a way may give you ideas for further analyses. As such, visualization tools are useful for exploratory analysis. In this lesson we will describe two different tools for visualization: a light-weight command-line based one and the Broad Institute’s Integrative Genomics Viewer (IGV) which requires software installation and transfer of files.
In order for us to visualize the alignment files, we will need to index the BAM file using samtools:
cd ~/dc_workshop
source $HOME/miniconda3/bin/activate
conda activate variant
samtools index results/bam/SRR2584866.aligned.sorted.bam5.8.1 Viewing with tview
Samtools implements a very simple text alignment viewer based on the GNU
ncurses library, called tview. This alignment viewer works with short indels and shows MAQ consensus.
It uses different colors to display mapping quality or base quality, subjected to users’ choice. Samtools viewer is known to work with a 130 GB alignment swiftly. Due to its text interface, displaying alignments over network is also very fast.
In order to visualize our mapped reads, we use tview, giving it the sorted bam file and the reference file:
cd ~/dc_workshop
source $HOME/miniconda3/bin/activate
conda activate variant
samtools tview results/bam/SRR2584866.aligned.sorted.bam data/ref_genome/ecoli_rel606.fastaThe first line of output shows the genome coordinates in our reference genome. The second line shows the reference
genome sequence. The third line shows the consensus sequence determined from the sequence reads. A . indicates
a match to the reference sequence, so we can see that the consensus from our sample matches the reference in most
locations. That is good! If that was not the case, we should probably reconsider our choice of reference.
Below the horizontal line, we can see all of the reads in our sample aligned with the reference genome. Only
positions where the called base differs from the reference are shown. You can use the arrow keys on your keyboard
to scroll or type ? for a help menu. To navigate to a specific position, type g. A dialogue box will appear. In
this box, type the name of the “chromosome” followed by a colon and the position of the variant you would like to view
(e.g. for this sample, type CP000819.1:50 to view the 50th base. Type Ctrl^C or q to exit tview.
5.9 Exercise
Visualize the alignment of the reads for our SRR2584866 sample. What variant is present at
position 4377265? What is the canonical nucleotide in that position?
5.10 Solution
CP000819.1:4,377,255-4,377,375
5.10.1 Viewing with IGV
IGV is a stand-alone browser, which has the advantage of being installed locally and providing fast access. Web-based genome browsers, like Ensembl or the UCSC browser, are slower, but provide more functionality. They not only allow for more polished and flexible visualization, but also provide easy access to a wealth of annotations and external data sources. This makes it straightforward to relate your data with information about repeat regions, known genes, epigenetic features or areas of cross-species conservation, to name just a few.
In order to use IGV, we will need to transfer some files and create a new folder.
mkdir -p ~/dc_workshop/files_for_igv
cd ~/dc_workshop/files_for_igvNow we will transfer our files to that new directory.
cp ~/dc_workshop/results/bam/SRR2584866.aligned.sorted.bam ~/dc_workshop/files_for_igv
cp ~/dc_workshop/results/bam/SRR2584866.aligned.sorted.bam.bai ~/dc_workshop/files_for_igv
cp ~/dc_workshop/data/ref_genome/ecoli_rel606.fasta ~/dc_workshop/files_for_igv
cp ~/dc_workshop/results/vcf/SRR2584866_final_variants.vcf ~/dc_workshop/files_for_igvNext, we need to open the IGV software. If you have not done so already, you can download IGV from the Broad Institute’s software page, double-click the .zip file
to unzip it, and then drag the program into your Applications folder.
- Open IGV.
- Load our reference genome file (
ecoli_rel606.fasta) into IGV using the “Load Genomes from File…” option under the “Genomes” pull-down menu. - Load our BAM file (
SRR2584866.aligned.sorted.bam) using the “Load from File…” option under the “File” pull-down menu. - Do the same with our VCF file (
SRR2584866_final_variants.vcf).
Your IGV browser should look like the screenshot below:
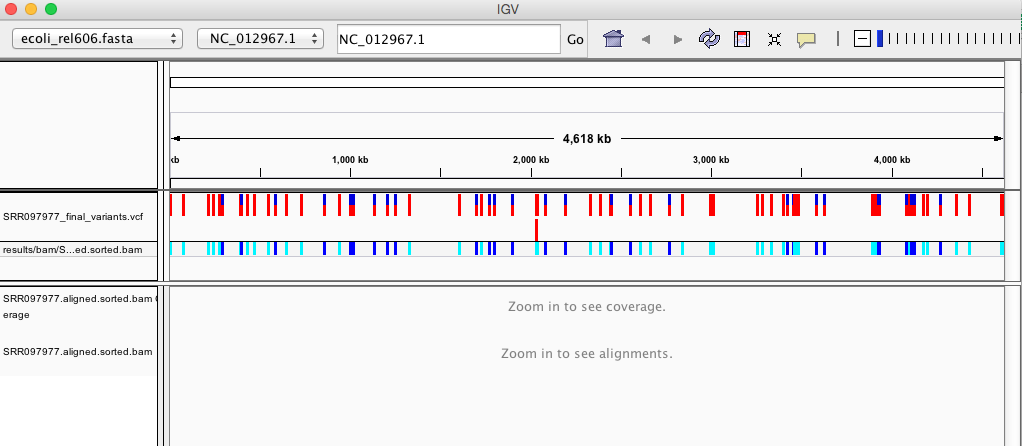
IGV
There should be two tracks: one coresponding to our BAM file and the other for our VCF file.
In the VCF track, each bar across the top of the plot shows the allele fraction for a single locus. The second bar shows the genotypes for each locus in each sample. We only have one sample called here, so we only see a single line. Dark blue = heterozygous, Cyan = homozygous variant, Grey = reference. Filtered entries are transparent.
Zoom in to inspect variants you see in your filtered VCF file to become more familiar with IGV. See how quality information corresponds to alignment information at those loci. Try an easy one like the variant present at position 4377265. Use this website and the links therein to understand how IGV colors the alignments.
Now that we have run through our workflow for a single sample, we want to repeat this workflow for our other five samples. However, we do not want to type each of these individual steps again five more times. That would be very time consuming and error-prone, and would become impossible as we gathered more and more samples. Luckily, we already know the tools we need to use to automate this workflow and run it on as many files as we want using a single line of code. Those tools are: wildcards, for loops, and bash scripts. We will use all three in the next lesson.
5.11 BWA alignment options
BWA consists of three algorithms: BWA-backtrack, BWA-SW and BWA-MEM. The first algorithm is designed for Illumina sequence reads up to 100bp, while the other two are for sequences ranging from 70bp to 1Mbp. BWA-MEM and BWA-SW share similar features such as long-read support and split alignment, but BWA-MEM, which is the latest, is generally recommended for high-quality queries as it is faster and more accurate.