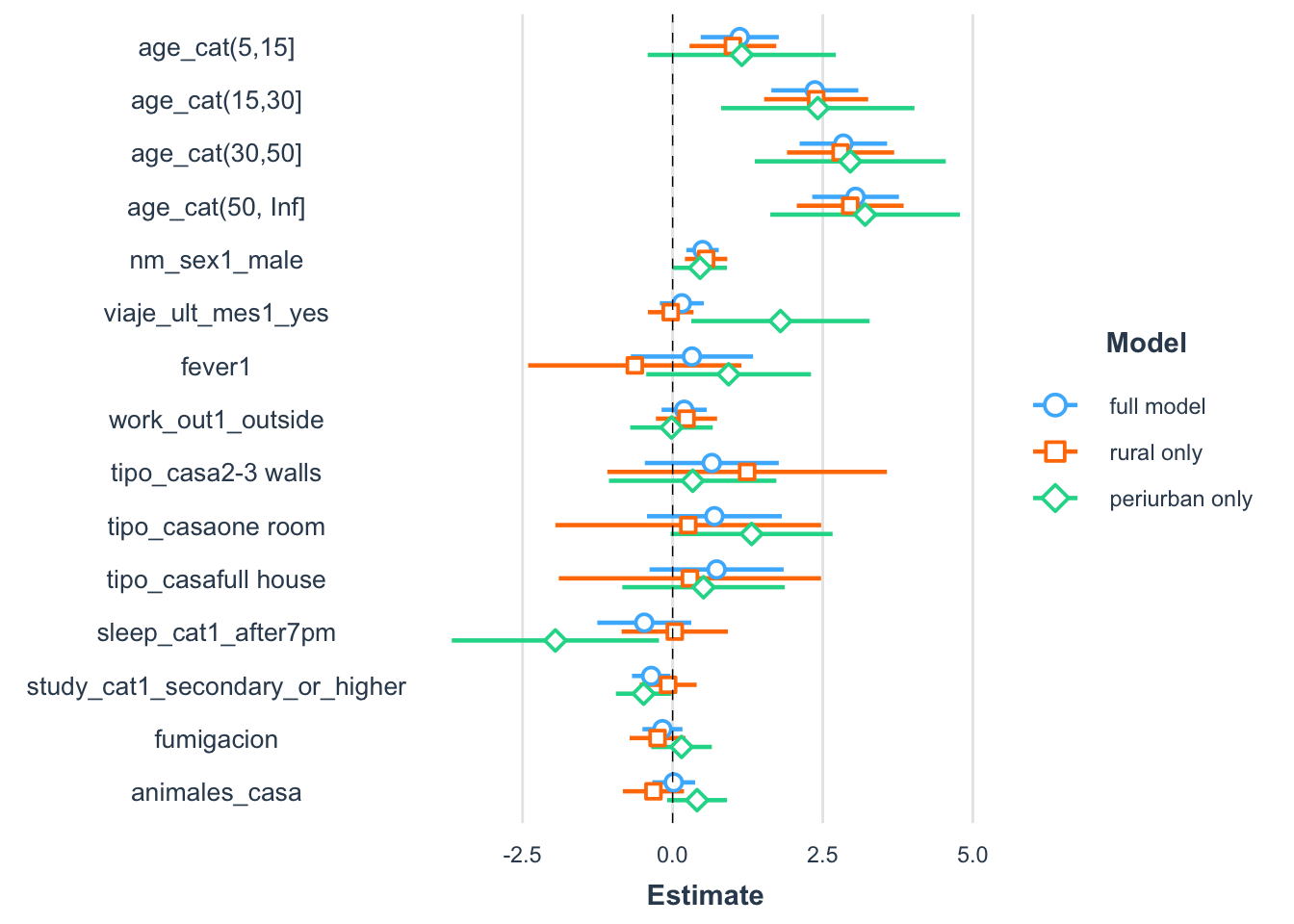
library(tidyverse)
library(sf)
dat <- d2 %>%
dplyr::select(id_house, comm, id_study:viaje_ult_mes, main_act_ec:hour_sleep, SEROPOSITIVE, area,
age_cat, fever, TREATMENT) %>%
st_set_geometry(NULL) %>%
separate(hour_sleep, into = c("hh","mm","ss"), sep = ":") %>%
mutate(viaje_ult_mes = ifelse(viaje_ult_mes=="9999", NA, viaje_ult_mes),
viaje_ult_mes = factor(ifelse(viaje_ult_mes=="2", "1_yes", "0_no")),
SEROPOSITIVE = ifelse(SEROPOSITIVE=="Positive",1,0),
work_out = factor(ifelse(as.numeric(main_act_ec)>0 & as.numeric(main_act_ec)<6, "1_outside",
"0_inside")),
study = as.numeric(as.character(nm_level_study)),
study = ifelse(study==9999,NA,study),
study_cat = factor(ifelse(study<5,"0_primary_or_lower", "1_secondary_or_higher")),
sleep_clean = as.numeric(hh),
sleep_clean = ifelse(sleep_clean<18,NA,sleep_clean),
sleep_cat = factor(ifelse(sleep_clean<19,"0_before7pm","1_after7pm")),
tipo_casa = factor(tipo_casa, labels = c("no walls", "2-3 walls", "one room","full house"))
) %>%
filter(complete.cases(.))
#epiDisplay::tab1(dat$sleep_cat)
Multilevel model
library(tidyverse)
library(jtools)
library(lme4)
m1 <- glmer(SEROPOSITIVE ~ age_cat + nm_sex + viaje_ult_mes + fever + work_out + tipo_casa +
sleep_cat + study_cat + fumigacion + animales_casa + (1|comm/id_house),
data = dat, family = binomial)
summ(m1, exp = T, confint = T)
|
Observations
|
1612
|
|
Dependent variable
|
SEROPOSITIVE
|
|
Type
|
Mixed effects generalized linear model
|
|
Family
|
binomial
|
|
Link
|
logit
|
|
AIC
|
1701.18
|
|
BIC
|
1798.11
|
|
Pseudo-R² (fixed effects)
|
0.20
|
|
Pseudo-R² (total)
|
0.43
|
|
Fixed Effects
|
|
|
exp(Est.)
|
2.5%
|
97.5%
|
z val.
|
p
|
|
(Intercept)
|
0.04
|
0.01
|
0.21
|
-3.91
|
0.00
|
|
age_cat(5,15]
|
3.06
|
1.60
|
5.86
|
3.37
|
0.00
|
|
age_cat(15,30]
|
10.68
|
5.18
|
22.04
|
6.41
|
0.00
|
|
age_cat(30,50]
|
17.17
|
8.29
|
35.57
|
7.65
|
0.00
|
|
age_cat(50, Inf]
|
21.04
|
10.22
|
43.32
|
8.27
|
0.00
|
|
nm_sex1_male
|
1.65
|
1.26
|
2.16
|
3.63
|
0.00
|
|
viaje_ult_mes1_yes
|
1.17
|
0.81
|
1.68
|
0.82
|
0.41
|
|
fever1
|
1.38
|
0.50
|
3.82
|
0.62
|
0.54
|
|
work_out1_outside
|
1.21
|
0.83
|
1.77
|
1.00
|
0.32
|
|
tipo_casa2-3 walls
|
1.92
|
0.63
|
5.86
|
1.15
|
0.25
|
|
tipo_casaone room
|
2.00
|
0.65
|
6.16
|
1.21
|
0.22
|
|
tipo_casafull house
|
2.08
|
0.68
|
6.35
|
1.28
|
0.20
|
|
sleep_cat1_after7pm
|
0.62
|
0.28
|
1.36
|
-1.18
|
0.24
|
|
study_cat1_secondary_or_higher
|
0.70
|
0.51
|
0.96
|
-2.20
|
0.03
|
|
fumigacion
|
0.84
|
0.60
|
1.18
|
-1.00
|
0.32
|
|
animales_casa
|
1.02
|
0.71
|
1.45
|
0.10
|
0.92
|
|
Random Effects
|
|
Group
|
Parameter
|
Std. Dev.
|
|
id_house:comm
|
(Intercept)
|
0.63
|
|
comm
|
(Intercept)
|
0.94
|
|
Grouping Variables
|
|
Group
|
# groups
|
ICC
|
|
id_house:comm
|
558
|
0.09
|
|
comm
|
10
|
0.19
|
m2 <- glmer(SEROPOSITIVE ~ age_cat + nm_sex + viaje_ult_mes + work_out + area +
study_cat + (1|comm/id_house),
data = dat, family = binomial)
summ(m2, exp = T, confint = T)
|
Observations
|
1612
|
|
Dependent variable
|
SEROPOSITIVE
|
|
Type
|
Mixed effects generalized linear model
|
|
Family
|
binomial
|
|
Link
|
logit
|
|
AIC
|
1690.04
|
|
BIC
|
1754.67
|
|
Pseudo-R² (fixed effects)
|
0.25
|
|
Pseudo-R² (total)
|
0.44
|
|
Fixed Effects
|
|
|
exp(Est.)
|
2.5%
|
97.5%
|
z val.
|
p
|
|
(Intercept)
|
0.02
|
0.01
|
0.08
|
-6.39
|
0.00
|
|
age_cat(5,15]
|
2.99
|
1.57
|
5.71
|
3.33
|
0.00
|
|
age_cat(15,30]
|
10.28
|
5.01
|
21.09
|
6.35
|
0.00
|
|
age_cat(30,50]
|
16.70
|
8.11
|
34.38
|
7.64
|
0.00
|
|
age_cat(50, Inf]
|
20.59
|
10.06
|
42.14
|
8.28
|
0.00
|
|
nm_sex1_male
|
1.66
|
1.27
|
2.17
|
3.69
|
0.00
|
|
viaje_ult_mes1_yes
|
1.15
|
0.79
|
1.65
|
0.73
|
0.47
|
|
work_out1_outside
|
1.16
|
0.80
|
1.69
|
0.77
|
0.44
|
|
area1_rural
|
3.13
|
1.01
|
9.70
|
1.98
|
0.05
|
|
study_cat1_secondary_or_higher
|
0.73
|
0.53
|
1.00
|
-1.98
|
0.05
|
|
Random Effects
|
|
Group
|
Parameter
|
Std. Dev.
|
|
id_house:comm
|
(Intercept)
|
0.66
|
|
comm
|
(Intercept)
|
0.79
|
|
Grouping Variables
|
|
Group
|
# groups
|
ICC
|
|
id_house:comm
|
558
|
0.10
|
|
comm
|
10
|
0.14
|
Model for rural areas (ICEMR)
dat_r <- dat %>%
filter(area == "1_rural")
m_rural <- glmer(SEROPOSITIVE ~ age_cat + nm_sex + viaje_ult_mes + fever + work_out + tipo_casa +
sleep_cat + study_cat + fumigacion + animales_casa + (1|comm/id_house),
data = dat_r, family = binomial)
summ(m_rural, exp = T, confint = T)
|
Observations
|
849
|
|
Dependent variable
|
SEROPOSITIVE
|
|
Type
|
Mixed effects generalized linear model
|
|
Family
|
binomial
|
|
Link
|
logit
|
|
AIC
|
960.16
|
|
BIC
|
1045.55
|
|
Pseudo-R² (fixed effects)
|
0.23
|
|
Pseudo-R² (total)
|
0.44
|
|
Fixed Effects
|
|
|
exp(Est.)
|
2.5%
|
97.5%
|
z val.
|
p
|
|
(Intercept)
|
0.06
|
0.00
|
0.79
|
-2.14
|
0.03
|
|
age_cat(5,15]
|
2.73
|
1.32
|
5.62
|
2.72
|
0.01
|
|
age_cat(15,30]
|
10.91
|
4.59
|
25.96
|
5.41
|
0.00
|
|
age_cat(30,50]
|
16.39
|
6.71
|
40.03
|
6.14
|
0.00
|
|
age_cat(50, Inf]
|
19.25
|
7.91
|
46.87
|
6.51
|
0.00
|
|
nm_sex1_male
|
1.75
|
1.23
|
2.48
|
3.09
|
0.00
|
|
viaje_ult_mes1_yes
|
0.97
|
0.66
|
1.42
|
-0.16
|
0.87
|
|
fever1
|
0.53
|
0.09
|
3.15
|
-0.70
|
0.49
|
|
work_out1_outside
|
1.26
|
0.76
|
2.10
|
0.89
|
0.37
|
|
tipo_casa2-3 walls
|
3.46
|
0.34
|
35.47
|
1.04
|
0.30
|
|
tipo_casaone room
|
1.30
|
0.14
|
11.87
|
0.23
|
0.82
|
|
tipo_casafull house
|
1.33
|
0.15
|
11.84
|
0.26
|
0.80
|
|
sleep_cat1_after7pm
|
1.04
|
0.43
|
2.51
|
0.08
|
0.94
|
|
study_cat1_secondary_or_higher
|
0.93
|
0.58
|
1.49
|
-0.31
|
0.76
|
|
fumigacion
|
0.78
|
0.49
|
1.24
|
-1.06
|
0.29
|
|
animales_casa
|
0.73
|
0.44
|
1.21
|
-1.23
|
0.22
|
|
Random Effects
|
|
Group
|
Parameter
|
Std. Dev.
|
|
id_house:comm
|
(Intercept)
|
0.54
|
|
comm
|
(Intercept)
|
0.96
|
|
Grouping Variables
|
|
Group
|
# groups
|
ICC
|
|
id_house:comm
|
245
|
0.07
|
|
comm
|
7
|
0.20
|
m_rural_2 <- glmer(SEROPOSITIVE ~ age_cat + nm_sex + fever + work_out + tipo_casa +
animales_casa + (1|comm/id_house),
data = dat_r, family = binomial)
summ(m_rural_2, exp = T, confint = T)
|
Observations
|
849
|
|
Dependent variable
|
SEROPOSITIVE
|
|
Type
|
Mixed effects generalized linear model
|
|
Family
|
binomial
|
|
Link
|
logit
|
|
AIC
|
953.43
|
|
BIC
|
1019.84
|
|
Pseudo-R² (fixed effects)
|
0.23
|
|
Pseudo-R² (total)
|
0.44
|
|
Fixed Effects
|
|
|
exp(Est.)
|
2.5%
|
97.5%
|
z val.
|
p
|
|
(Intercept)
|
0.06
|
0.01
|
0.67
|
-2.29
|
0.02
|
|
age_cat(5,15]
|
2.73
|
1.33
|
5.63
|
2.73
|
0.01
|
|
age_cat(15,30]
|
10.66
|
4.58
|
24.81
|
5.49
|
0.00
|
|
age_cat(30,50]
|
16.06
|
6.65
|
38.82
|
6.17
|
0.00
|
|
age_cat(50, Inf]
|
18.95
|
7.79
|
46.13
|
6.48
|
0.00
|
|
nm_sex1_male
|
1.74
|
1.23
|
2.46
|
3.11
|
0.00
|
|
fever1
|
0.54
|
0.09
|
3.19
|
-0.68
|
0.50
|
|
work_out1_outside
|
1.27
|
0.77
|
2.11
|
0.93
|
0.35
|
|
tipo_casa2-3 walls
|
3.38
|
0.33
|
34.47
|
1.03
|
0.30
|
|
tipo_casaone room
|
1.24
|
0.14
|
11.24
|
0.19
|
0.85
|
|
tipo_casafull house
|
1.33
|
0.15
|
11.75
|
0.25
|
0.80
|
|
animales_casa
|
0.68
|
0.42
|
1.11
|
-1.54
|
0.12
|
|
Random Effects
|
|
Group
|
Parameter
|
Std. Dev.
|
|
id_house:comm
|
(Intercept)
|
0.57
|
|
comm
|
(Intercept)
|
0.95
|
|
Grouping Variables
|
|
Group
|
# groups
|
ICC
|
|
id_house:comm
|
245
|
0.07
|
|
comm
|
7
|
0.20
|
Model for peri-urban areas (CAM)
dat_p <- dat %>%
filter(area == "0_periurban")
m_peri <- glmer(SEROPOSITIVE ~ age_cat + nm_sex + viaje_ult_mes + fever + work_out + tipo_casa +
sleep_cat + study_cat + fumigacion + animales_casa + (1|comm/id_house),
data = dat_p, family = binomial)
summ(m_peri, exp = T, confint = T)
|
Observations
|
763
|
|
Dependent variable
|
SEROPOSITIVE
|
|
Type
|
Mixed effects generalized linear model
|
|
Family
|
binomial
|
|
Link
|
logit
|
|
AIC
|
737.17
|
|
BIC
|
820.64
|
|
Pseudo-R² (fixed effects)
|
0.25
|
|
Pseudo-R² (total)
|
0.36
|
|
Fixed Effects
|
|
|
exp(Est.)
|
2.5%
|
97.5%
|
z val.
|
p
|
|
(Intercept)
|
0.06
|
0.00
|
0.74
|
-2.18
|
0.03
|
|
age_cat(5,15]
|
3.16
|
0.66
|
15.16
|
1.44
|
0.15
|
|
age_cat(15,30]
|
11.22
|
2.24
|
56.14
|
2.94
|
0.00
|
|
age_cat(30,50]
|
19.27
|
3.93
|
94.54
|
3.65
|
0.00
|
|
age_cat(50, Inf]
|
24.68
|
5.09
|
119.71
|
3.98
|
0.00
|
|
nm_sex1_male
|
1.58
|
1.01
|
2.47
|
2.00
|
0.05
|
|
viaje_ult_mes1_yes
|
6.02
|
1.36
|
26.58
|
2.37
|
0.02
|
|
fever1
|
2.54
|
0.64
|
10.02
|
1.33
|
0.18
|
|
work_out1_outside
|
0.98
|
0.49
|
1.95
|
-0.05
|
0.96
|
|
tipo_casa2-3 walls
|
1.40
|
0.35
|
5.63
|
0.47
|
0.64
|
|
tipo_casaone room
|
3.73
|
0.97
|
14.35
|
1.91
|
0.06
|
|
tipo_casafull house
|
1.67
|
0.43
|
6.46
|
0.74
|
0.46
|
|
sleep_cat1_after7pm
|
0.14
|
0.03
|
0.80
|
-2.22
|
0.03
|
|
study_cat1_secondary_or_higher
|
0.62
|
0.39
|
0.98
|
-2.05
|
0.04
|
|
fumigacion
|
1.16
|
0.70
|
1.92
|
0.57
|
0.57
|
|
animales_casa
|
1.50
|
0.91
|
2.47
|
1.60
|
0.11
|
|
Random Effects
|
|
Group
|
Parameter
|
Std. Dev.
|
|
id_house:comm
|
(Intercept)
|
0.75
|
|
comm
|
(Intercept)
|
0.00
|
|
Grouping Variables
|
|
Group
|
# groups
|
ICC
|
|
id_house:comm
|
313
|
0.15
|
|
comm
|
3
|
0.00
|
m_peri_2 <- glmer(SEROPOSITIVE ~ age_cat + nm_sex +
study_cat + (1|comm/id_house),
data = dat_p, family = binomial)
summ(m_peri_2, exp = T, confint = T)
|
Observations
|
763
|
|
Dependent variable
|
SEROPOSITIVE
|
|
Type
|
Mixed effects generalized linear model
|
|
Family
|
binomial
|
|
Link
|
logit
|
|
AIC
|
743.84
|
|
BIC
|
785.57
|
|
Pseudo-R² (fixed effects)
|
0.18
|
|
Pseudo-R² (total)
|
0.33
|
|
Fixed Effects
|
|
|
exp(Est.)
|
2.5%
|
97.5%
|
z val.
|
p
|
|
(Intercept)
|
0.03
|
0.01
|
0.12
|
-4.68
|
0.00
|
|
age_cat(5,15]
|
3.14
|
0.68
|
14.58
|
1.46
|
0.14
|
|
age_cat(15,30]
|
10.12
|
2.10
|
48.82
|
2.88
|
0.00
|
|
age_cat(30,50]
|
18.01
|
3.84
|
84.43
|
3.67
|
0.00
|
|
age_cat(50, Inf]
|
23.66
|
5.13
|
109.15
|
4.06
|
0.00
|
|
nm_sex1_male
|
1.58
|
1.05
|
2.38
|
2.19
|
0.03
|
|
study_cat1_secondary_or_higher
|
0.60
|
0.38
|
0.96
|
-2.14
|
0.03
|
|
Random Effects
|
|
Group
|
Parameter
|
Std. Dev.
|
|
id_house:comm
|
(Intercept)
|
0.81
|
|
comm
|
(Intercept)
|
0.26
|
|
Grouping Variables
|
|
Group
|
# groups
|
ICC
|
|
id_house:comm
|
313
|
0.16
|
|
comm
|
3
|
0.02
|