Chapter 2 Aim1: PREDICTION CAPACITY [Random Effects model]
2.1 Data
rm(list=ls())
library(raster)
library(sf)
library(rasterVis)
library(tidyverse)
library(haven)
library(stringr)
library(scales)
library(cowplot)
library(ggpubr)dat <- readRDS(file.path(wd, "/dat_NL_MAL.RData")) %>%
filter(nrohab>20) %>%
mutate(fal_rate = 100*fal/nrohab,
viv_rate = 100*viv/nrohab)
dat_year <- dat %>%
ungroup() %>%
group_by(year, id_loc) %>%
filter(nrohab>20) %>%
summarise(provinc = first(province.x),
fal = sum(fal),
viv = sum(viv),
nrohab = mean(nrohab),
MEI_ike_udel = mean(MEI_ike_udel),
nl_viirs_2 = mean(nl_viirs_2, na.rm=T),
nl_viirs_2_max = max(nl_viirs_2, na.rm=T)) %>%
mutate(fal_rate = 1000*fal/nrohab,
viv_rate = 1000*viv/nrohab,
bin_viv = ifelse(viv>0, 1,0),
bin_fal = ifelse(fal>0, 1,0))
dat_year_2 <- dat_year %>%
filter(nl_viirs_2_max < quantile(dat_year$nl_viirs_2_max, 0.99),
!is.na(nl_viirs_2)) %>%
mutate(nl_viirs_2_max_sq = nl_viirs_2_max^2,
drop = ifelse(viv>nrohab,1,0),
drop = ifelse(fal>nrohab,1,drop)) %>%
ungroup() %>%
group_by(id_loc) %>%
mutate(drop2 = max(drop, na.rm = T)) %>%
filter(drop==0)
db <- readRDS("~/Dropbox/Work/SESYNC/Projects/Malaria/Village Level [Javier]/Analysis/output/data_sf.RData") %>%
mutate(id2 = row_number())
villages <- db %>%
dplyr::select(id_loc, id2) %>%
inner_join(st_set_geometry(db, NULL) %>%
dplyr::select(id_loc, nrohab, village.x, commune, province.x, uid, id2) %>%
distinct(id_loc, .keep_all = T), by=c("id_loc", "id2")) %>%
filter(id_loc!="")
river <- st_read("~/Dropbox/Resources/GIS/Hidro_HDX/rionavegables/Rio_navegables.shp") %>%
filter(i==1) %>%
st_transform("+proj=longlat +datum=WGS84 +no_defs")
d <- read_csv("~/Dropbox/Work/SESYNC/Projects/Malaria/Village Level [Javier]/Analysis/output/dist_amazon.csv") %>%
dplyr::select(id_loc, distance) %>%
mutate(distance = distance*111111)
villages <- villages %>%
inner_join(d, by = "id_loc")
# NOT RUN
a <- dat_year_2 %>%
distinct(id_loc, .keep_all = T)
rm(a)2.2 Model
library(lme4)
library(skimr)
library(broom.mixed)
fit1 <- glmer.nb(viv ~ nl_viirs_2_max + (1 + nl_viirs_2_max | id_loc) + offset(log(nrohab)), dat_year_2)
coeff1 <- ranef(fit1) %>%
augment() %>%
mutate(sig = ifelse(lb<0 & ub>0,0,1),
nl_viirs_2_max = estimate,
id_loc = as.character(level))
coeff1_s <- coeff1 %>%
filter(sig==1 & variable == "nl_viirs_2_max") %>%
select(id_loc, nl_viirs_2_max)
coeff1 <- coeff1 %>%
filter(variable == "nl_viirs_2_max") %>%
select(id_loc, nl_viirs_2_max)
skim(coeff1, nl_viirs_2_max)
fit3 <- glmer.nb(viv ~ nl_viirs_2_max + provinc + (1 + nl_viirs_2_max | id_loc) + offset(log(nrohab)), dat_year_2)
coeff3 <- ranef(fit3) %>%
augment() %>%
mutate(sig = ifelse(lb<0 & ub>0,0,1),
nl_viirs_2_max2 = estimate,
id_loc = as.character(level))
coeff3_s <- coeff3 %>%
filter(sig==1 & variable == "nl_viirs_2_max") %>%
select(id_loc, nl_viirs_2_max2)
coeff3 <- coeff3 %>%
filter(variable == "nl_viirs_2_max") %>%
select(id_loc, nl_viirs_2_max2)
skim(coeff3, nl_viirs_2_max2)2.3 Model sumamry
library(jtools)
summ(fit1)| Observations | 10629 |
| Dependent variable | viv |
| Type | Mixed effects generalized linear model |
| Family | Negative Binomial(0.6838) |
| Link | log |
| AIC | 51608.00 |
| BIC | 51651.63 |
| Est. | S.E. | z val. | p | |
|---|---|---|---|---|
| (Intercept) | -4.05 | 0.06 | -73.40 | 0.00 |
| nl_viirs_2_max | -0.21 | 0.11 | -1.88 | 0.06 |
| Group | Parameter | Std. Dev. |
|---|---|---|
| id_loc | (Intercept) | 1.99 |
| id_loc | nl_viirs_2_max | 0.17 |
| Group | # groups | ICC |
|---|---|---|
| id_loc | 1555 | 0.38 |
summ(fit3)| Observations | 10629 |
| Dependent variable | viv |
| Type | Mixed effects generalized linear model |
| Family | Negative Binomial(0.6485) |
| Link | log |
| AIC | 51330.61 |
| BIC | 51425.13 |
| Est. | S.E. | z val. | p | |
|---|---|---|---|---|
| (Intercept) | -5.22 | 0.14 | -36.01 | 0.00 |
| nl_viirs_2_max | -0.34 | 0.14 | -2.47 | 0.01 |
| provincDATEM DEL MARA_ON | 1.60 | 0.19 | 8.38 | 0.00 |
| provincLORETO | 1.95 | 0.19 | 10.14 | 0.00 |
| provincMARISCAL RAMON CASTILLA | 1.66 | 0.21 | 7.92 | 0.00 |
| provincMAYNAS | 1.55 | 0.17 | 9.13 | 0.00 |
| provincPUTUMAYO | 0.50 | 0.31 | 1.58 | 0.11 |
| provincREQUENA | -0.39 | 0.23 | -1.69 | 0.09 |
| provincUCAYALI | -3.10 | 0.48 | -6.44 | 0.00 |
| Group | Parameter | Std. Dev. |
|---|---|---|
| id_loc | (Intercept) | 1.77 |
| id_loc | nl_viirs_2_max | 0.46 |
| Group | # groups | ICC |
|---|---|---|
| id_loc | 1555 | 0.23 |
2.4 Output
ggplot() +
geom_density(data = coeff1, aes(nl_viirs_2_max, fill = "viirs"), col = NA, alpha = .7) +
geom_density(data = coeff3, aes(nl_viirs_2_max2, fill = "viirs + province"), col = NA, alpha = .7) +
labs(fill = "Model", x = "viirs beta coefficients", y = "Density") +
scale_y_continuous(expand = c(0, 0)) +
scale_fill_manual(values = c("viirs" = "#9AC0CD", "viirs + province" = "#8B475D")) +
theme_bw()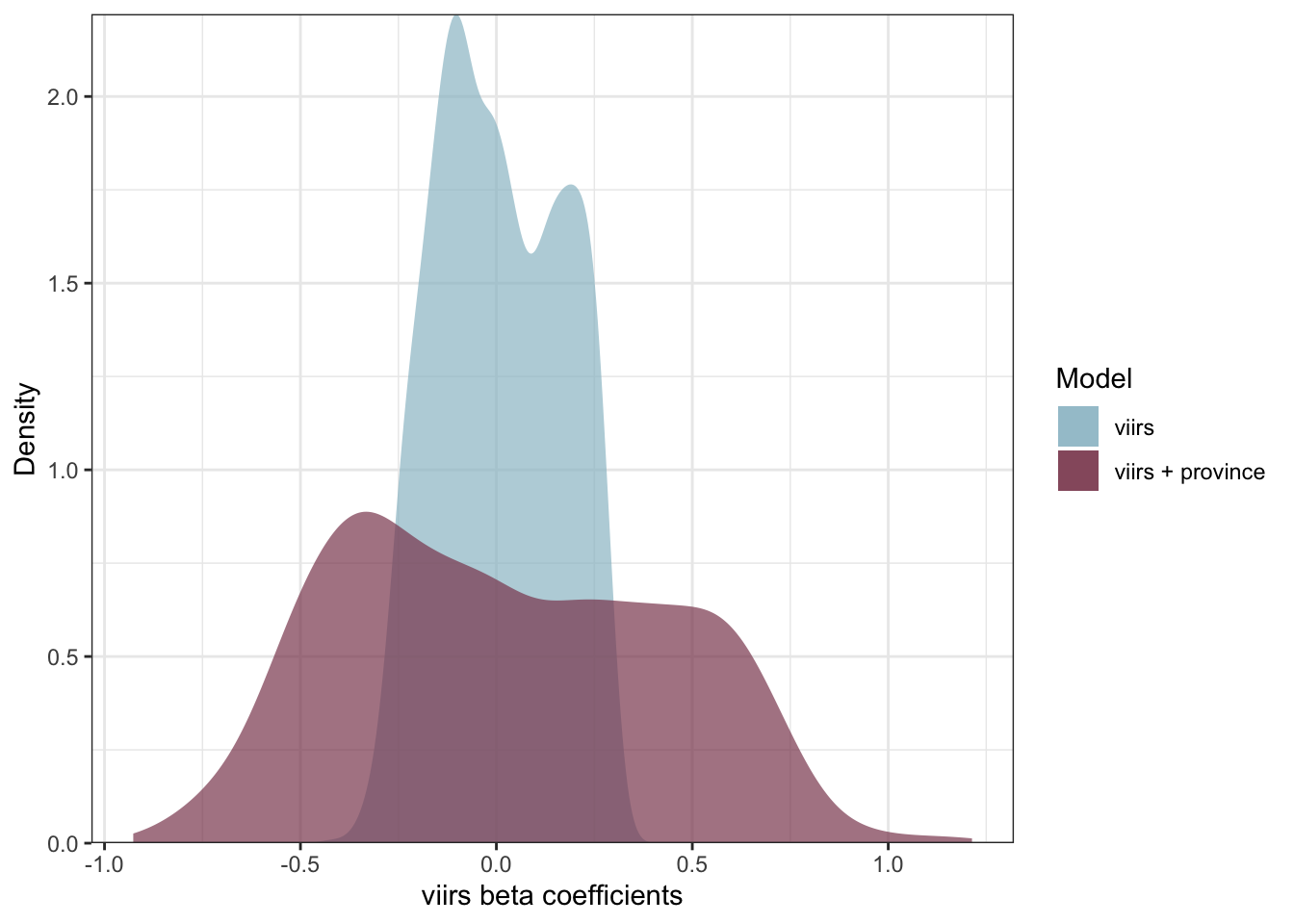
ggplot() +
geom_density(data = coeff1_s, aes(nl_viirs_2_max, fill = "viirs"), col = NA, alpha = .7) +
geom_density(data = coeff3_s, aes(nl_viirs_2_max2, fill = "viirs + province"), col = NA, alpha = .7) +
labs(fill = "Model", x = "viirs beta coefficients", y = "Density") +
scale_y_continuous(expand = c(0, 0)) +
scale_fill_manual(values = c("viirs" = "#9AC0CD", "viirs + province" = "#8B475D")) +
theme_bw()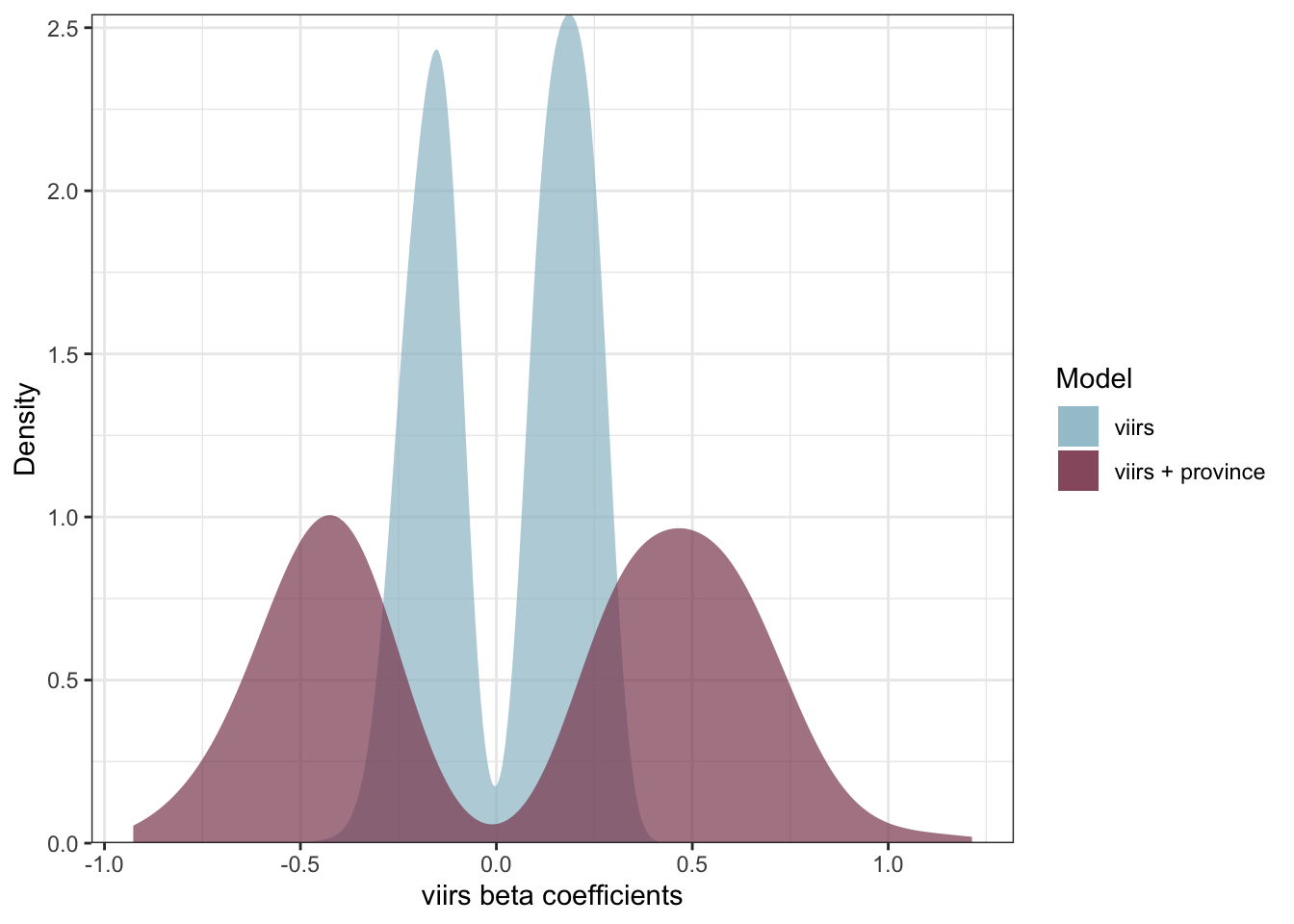
2.4.1 Spatial distribution
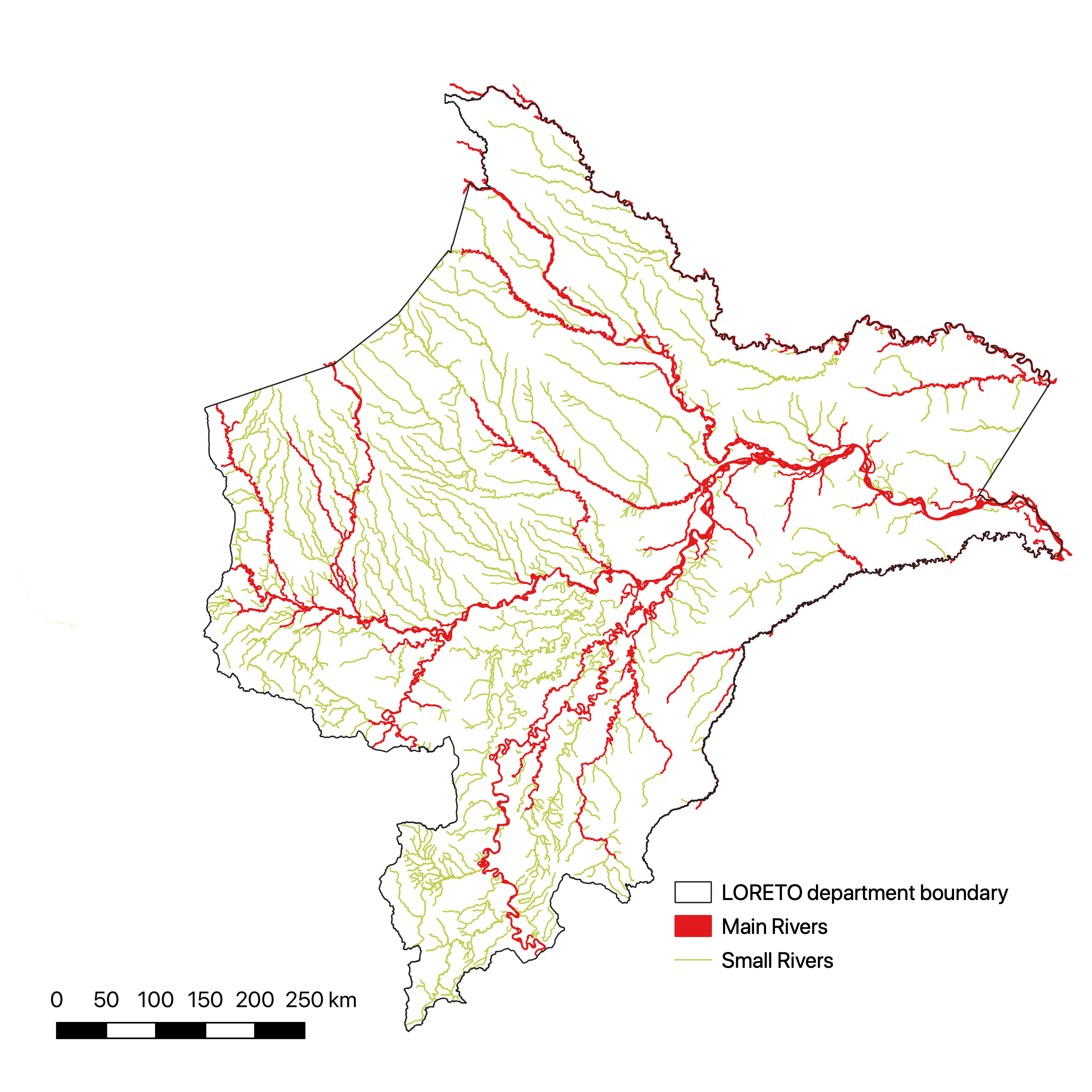
coeff3.sf <- villages %>%
inner_join(coeff3, by="id_loc") %>%
rename(NOMBPROV = province.x,
NOMBDIST = commune)
coeff3_s.sf <- villages %>%
inner_join(coeff3_s, by="id_loc") %>%
rename(NOMBPROV = province.x,
NOMBDIST = commune)
area.sf <- st_read("~/Dropbox/Work/Climate and Malaria/Analysis/Data/MapLOR_DIST/MapLOR_DIST.shp")## Reading layer `MapLOR_DIST' from data source `/Users/gcarrasco/Dropbox/Work/Climate and Malaria/Analysis/Data/MapLOR_DIST/MapLOR_DIST.shp' using driver `ESRI Shapefile'
## Simple feature collection with 49 features and 19 fields
## geometry type: POLYGON
## dimension: XY
## bbox: xmin: -77.82596 ymin: -8.716139 xmax: -69.94904 ymax: -0.03860597
## epsg (SRID): 4326
## proj4string: +proj=longlat +datum=WGS84 +no_defsl <- st_read("~/Dropbox/Resources/GIS/per_admbnda_adm1_2018/per_admbnda_adm1_2018.shp") %>%
filter(ADM1_ES =="Loreto")## Reading layer `per_admbnda_adm1_2018' from data source `/Users/gcarrasco/Dropbox/Resources/GIS/per_admbnda_adm1_2018/per_admbnda_adm1_2018.shp' using driver `ESRI Shapefile'
## Simple feature collection with 25 features and 7 fields
## geometry type: MULTIPOLYGON
## dimension: XY
## bbox: xmin: -81.32823 ymin: -18.35093 xmax: -68.65228 ymax: -0.03860597
## epsg (SRID): 4326
## proj4string: +proj=longlat +datum=WGS84 +no_defs#river
bb <- st_bbox(l)
bb[1] <- -77.6
river.l <- st_crop(river,bb)
area.sf_coeff_s <- area.sf %>%
inner_join(st_set_geometry(coeff3_s.sf, NULL) %>%
group_by(NOMBDIST) %>%
summarise(coeff = mean(nl_viirs_2_max2)),
by = "NOMBDIST")
area.sf_coeff <- area.sf %>%
inner_join(st_set_geometry(coeff3.sf, NULL) %>%
group_by(NOMBDIST) %>%
summarise(coeff = mean(nl_viirs_2_max2)),
by = "NOMBDIST")2.5 Figures
2.5.1 Figure post1
ggplot() +
geom_sf(data = l, size = 0.5, col = "black") +
geom_sf(data = river.l, col="#8B1C62", size = 0.3) +
geom_sf(data = coeff3_s.sf, aes(col=nl_viirs_2_max2, size = nrohab), alpha=.3) +
scale_color_gradient2(low = "#00688B", mid = "white", high = "#CD3700") +
labs(color = "viirs RE slope") +
theme_void() +
theme(axis.text = element_text(size = 15))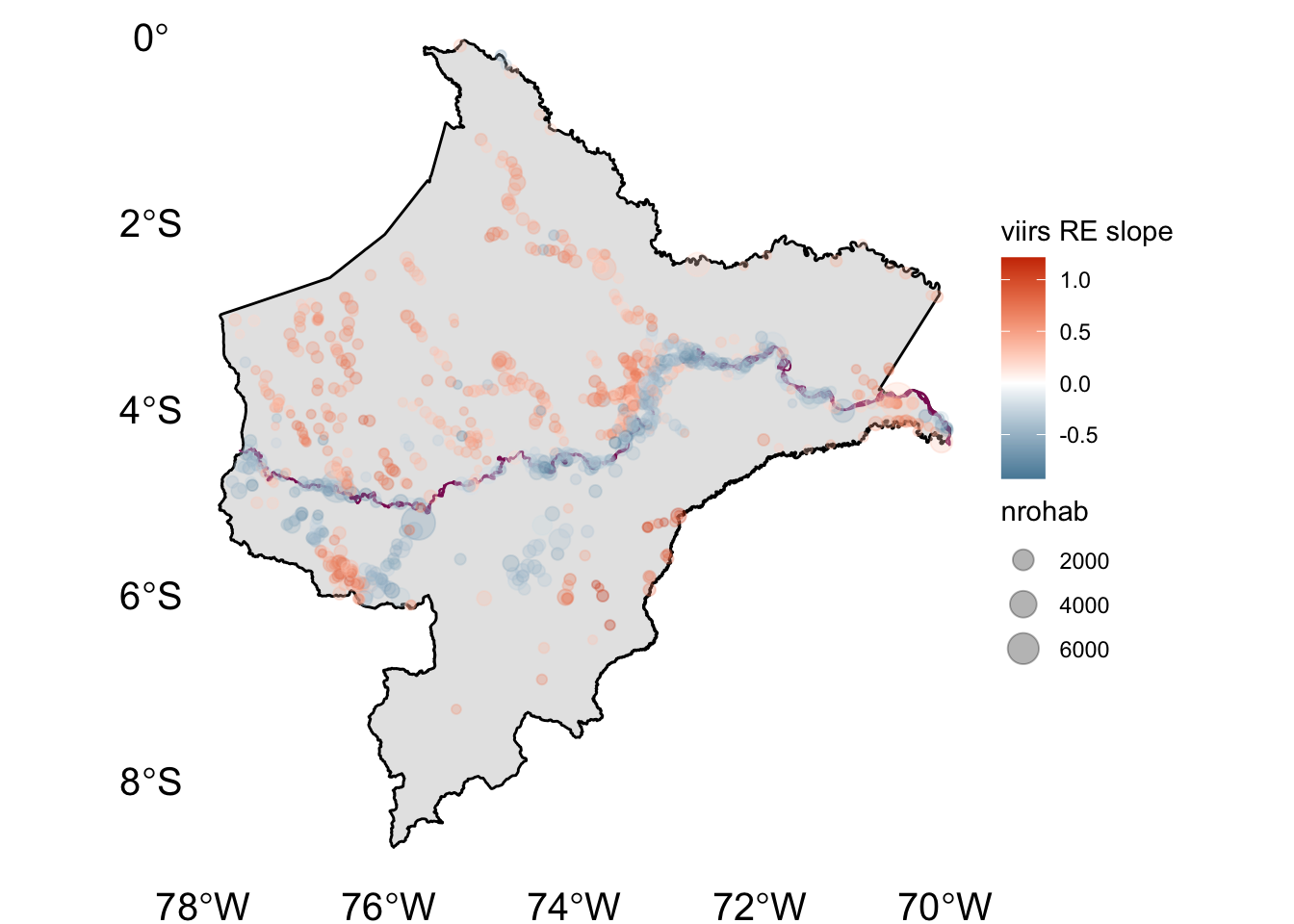
2.5.2 Figure post2
area.sf_top = coeff3_s.sf %>%
mutate(low = ifelse(ntile(nl_viirs_2_max2,100)<3,"low","a"),
high = ifelse(ntile(nl_viirs_2_max2,100)>98,"high","a"),
beta = ifelse(low=="a",high,low)) %>%
filter(beta!="a")
ggplot() +
geom_sf(data = l, fill=NA, size = 0.5, col = "black") +
geom_sf(data = river.l, col="#8B1C62", size = 0.3) +
geom_sf(data = area.sf_top,
aes(col=beta), size = 2, alpha=.5) +
labs(color = "viirs RE slope") +
theme_void() +
theme(axis.text = element_text(size = 15))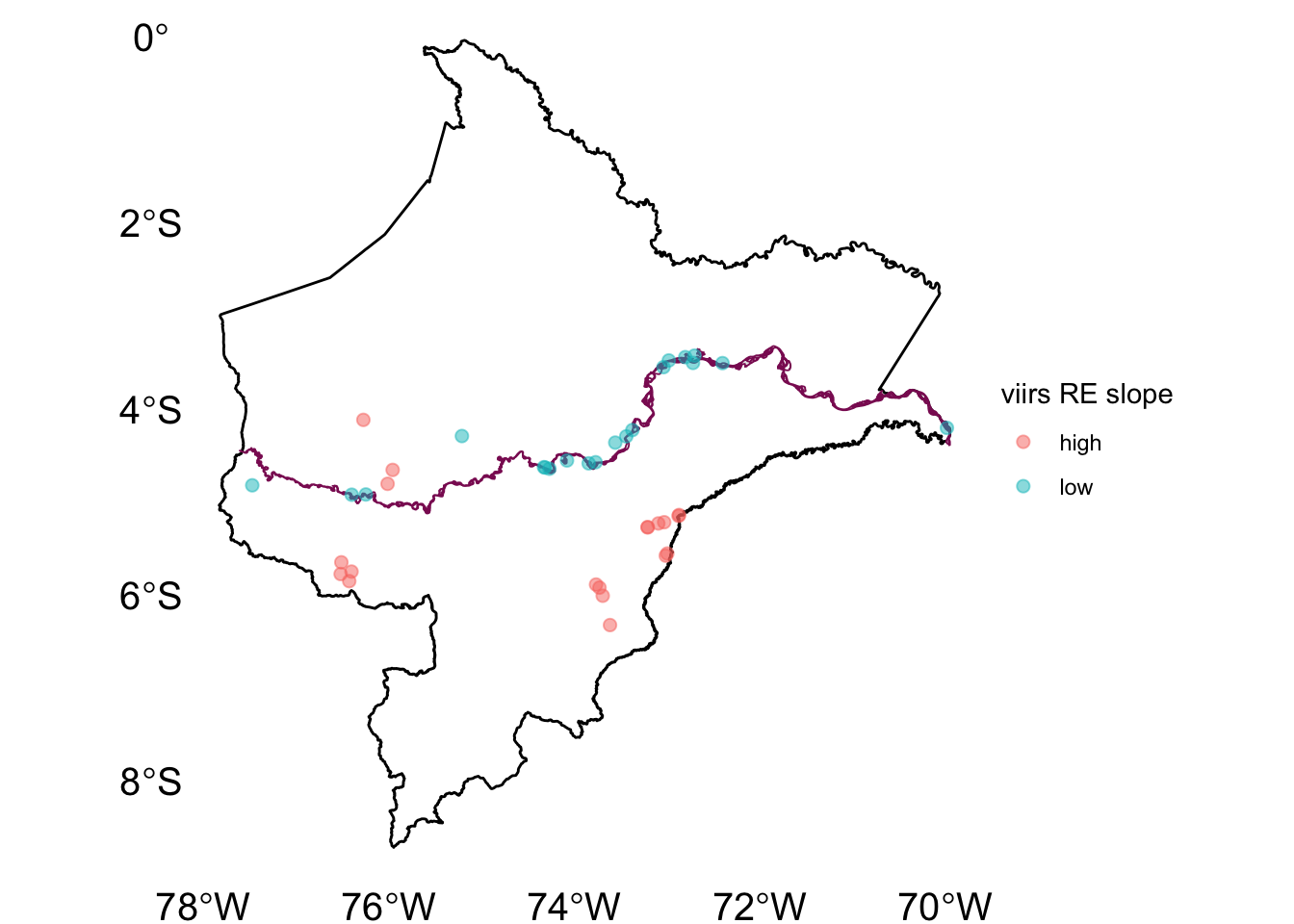
2.5.3 Figure post3 [skip]
# area.sf_top %>%
# filter(beta=="low") %>%
# st_set_geometry(NULL) %>%
# dplyr::select(id_loc) %>%
# left_join(dat_year_2, by = "id_loc") %>%
# ggplot(aes(x = year, group = id_loc)) +
# geom_line(aes(y = viv_rate, col = "viv rate")) +
# geom_line(aes(y = nl_viirs_2_max*10, col = "viirs")) +
# scale_color_manual(values = c("viv rate" = "black", "viirs" = "red")) +
# labs(col = "var", x = "year", y = "", title = "villages with most negative betas") +
# scale_y_continuous("viv rate", sec.axis = sec_axis(~ . / 10, name = "viirs")) +
# facet_wrap(.~id_loc, scales = "free_y")
#
# area.sf_top %>%
# filter(beta=="high") %>%
# st_set_geometry(NULL) %>%
# dplyr::select(id_loc) %>%
# left_join(dat_year_2, by = "id_loc") %>%
# ggplot(aes(x = year, group = id_loc)) +
# geom_line(aes(y = viv_rate, col = "viv rate")) +
# geom_line(aes(y = nl_viirs_2_max*3000, col = "viirs")) +
# scale_color_manual(values = c("viv rate" = "black", "viirs" = "red")) +
# labs(col = "var", x = "year", y = "", title = "villages with most positive betas") +
# scale_y_continuous("viv rate", sec.axis = sec_axis(~ . / 3000, name = "viirs")) +
# facet_wrap(.~id_loc, scales = "free_y")2.5.4 Figure post4 [Figure 2]
(map <- area.sf_coeff_s %>% #mutate(NOMBDIST2 = factor(NOMBDIST, levels=unique(NOMBDIST[order(NOMBPROV)]), ordered=TRUE)) %>%
ggplot() +
geom_sf(aes(fill=coeff), size = 0.5, col = NA) +
geom_sf(data = l, fill=NA, size = 0.5, col = "black") +
geom_sf(data = river.l, aes(color="Amazon River"), size = 0.3, show.legend = "line") +
scale_fill_gradient2(low = "#00688B", mid = "white", high = "#CD3700") +
scale_color_manual(values = "#8B1C62", name = "") +
labs(fill = "Mean Coeff") +
theme_bw() +
theme(axis.text = element_text(size = 10),
legend.position = "bottom"))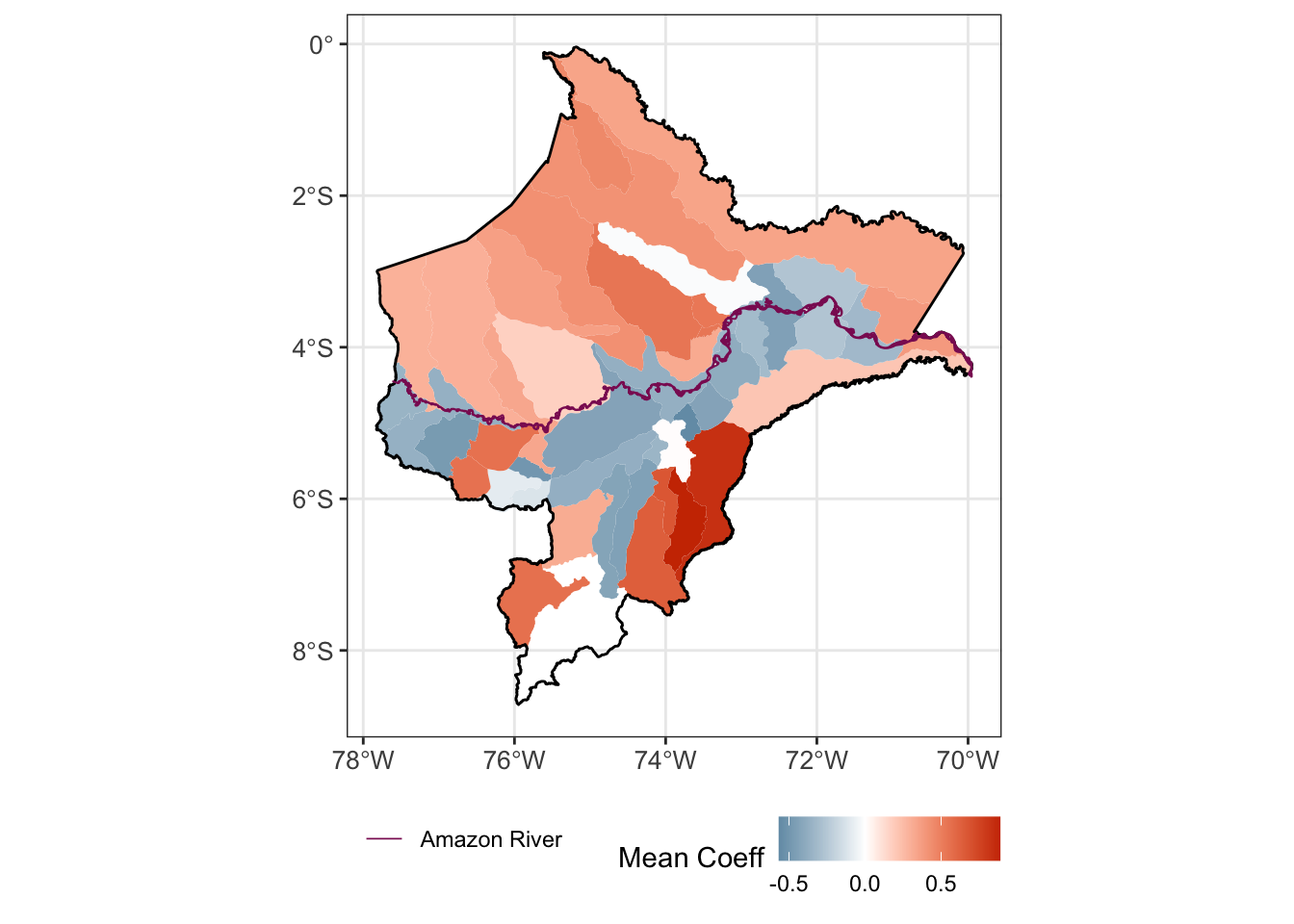
library(cowplot)
f2 <- plot_grid(map, labels = c("A"))
#ggsave(f2, filename = "./_out/f2.png", width = 5, height = 6, dpi = 300)library(ggExtra)
a <- dat %>%
filter(!is.na(nl_viirs_2)) %>%
st_set_geometry(NULL) %>%
group_by(id_loc) %>%
summarise(nl_viirs_2 = first(nl_viirs_2)) %>%
mutate(nl_viirs_2 = ifelse(nl_viirs_2<0,0,nl_viirs_2))
b <- coeff3.sf %>% #cambiar
inner_join(a, by= "id_loc") %>%
inner_join(area.sf_coeff %>% # cambiar
st_set_geometry(NULL) %>%
mutate(dist = ifelse(coeff<0, "main river", "small river")) %>%
dplyr::select(NOMBDIST, dist),
by= "NOMBDIST")2.5.5 VIIRS baseline vs. beta
b %>%
ggplot(aes(x=nl_viirs_2, y=nl_viirs_2_max2)) +
geom_point(alpha=.3) +
scale_x_continuous(trans="log10", labels = scales::comma_format()) +
labs(x = "viirs baseline", y = "viirs beta") +
geom_smooth(method = "loess", col="red", fill="red") +
stat_smooth(method = "lm", formula = y ~ poly(x, 2), col="blue", fill="blue") +
theme_bw()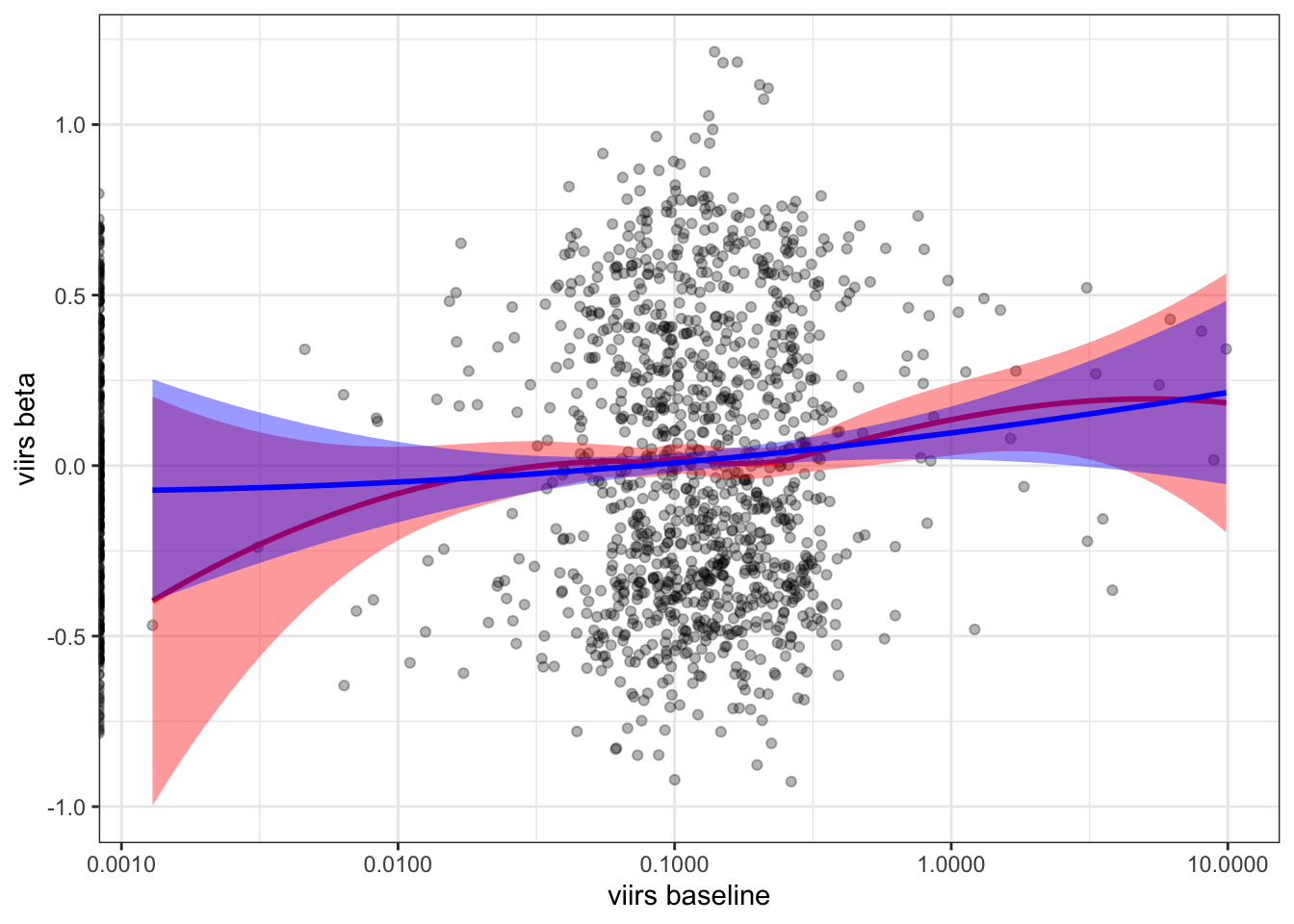
2.5.6 Distance to Amazon vs. beta [Fig 3a]
f3a.p <- b %>%
ggplot(aes(x=distance, y=nl_viirs_2_max2)) +
geom_point(alpha=.3, shape = 3) +
scale_x_continuous(trans="log10", labels = scales::comma_format()) +
labs(x = "Distance to Amazon River (m)", y = "VIIRS beta") +
#geom_smooth(method = "loess", col="red", fill="red") +
stat_smooth(method = "lm", formula = y ~ poly(x, 2), col="#FDE725FF", fill="#FDE725FF") +
theme_bw()
f3a <- ggMarginal(f3a.p, xparams = list(fill = "#481568FF", col = "#440154FF", alpha=.3), #margins = 'x',
yparams = list(fill = "#29AF7FFF", col = "#20A386FF", alpha=.3))
f3a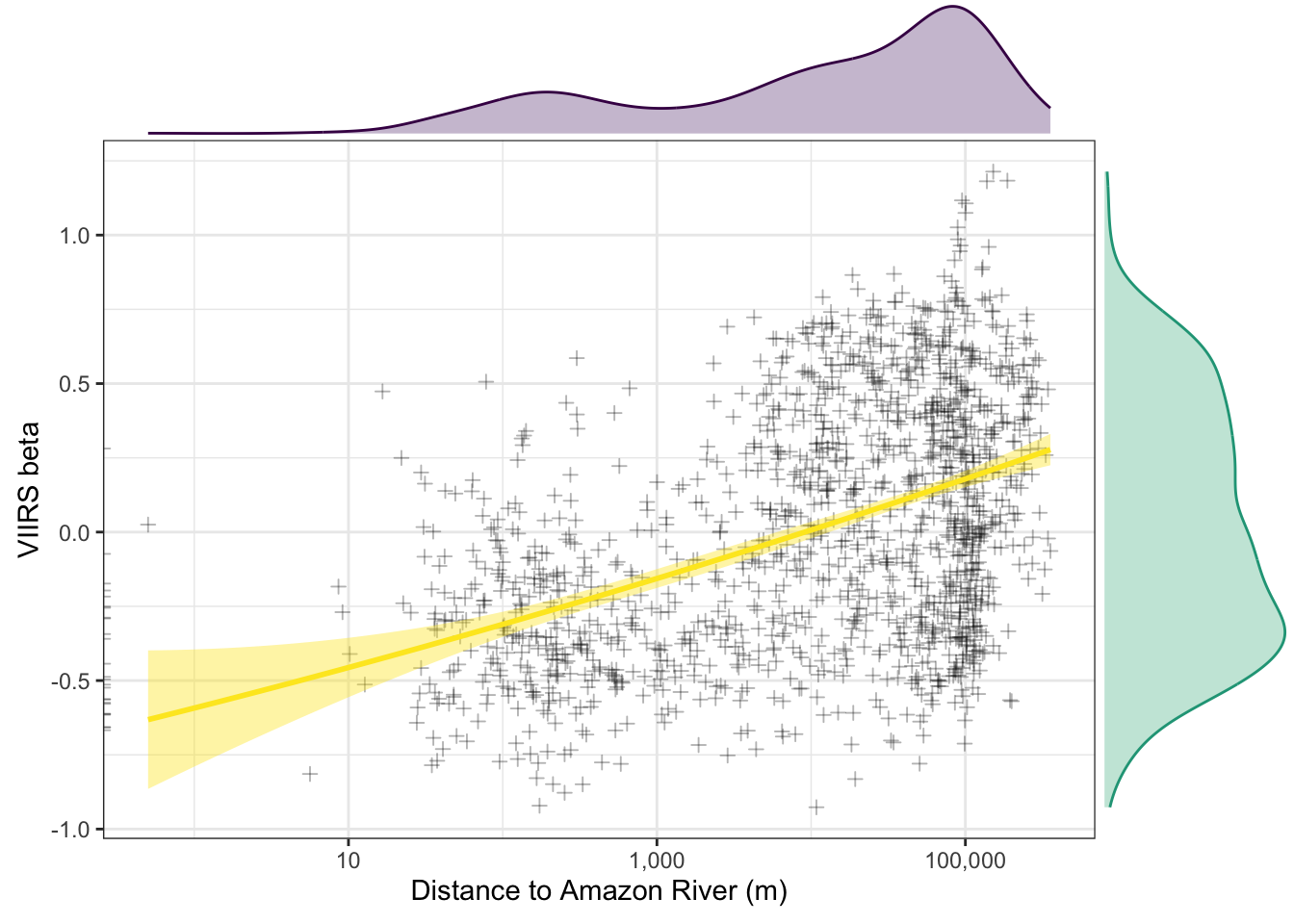
2.5.7 Distance to Amazon vs. VIIRS baseline
b %>%
ggplot(aes(x=distance, y=nl_viirs_2)) +
geom_point(alpha=.3) +
scale_x_continuous(trans="log10", labels = scales::comma_format()) +
scale_y_continuous(trans="log10", labels = scales::comma_format()) +
labs(x = "distance to Amazon river", y = "viirs baseline") +
geom_smooth(method = "loess", col="red", fill="red") +
stat_smooth(method = "lm", formula = y ~ poly(x, 2), col="blue", fill="blue") +
theme_bw()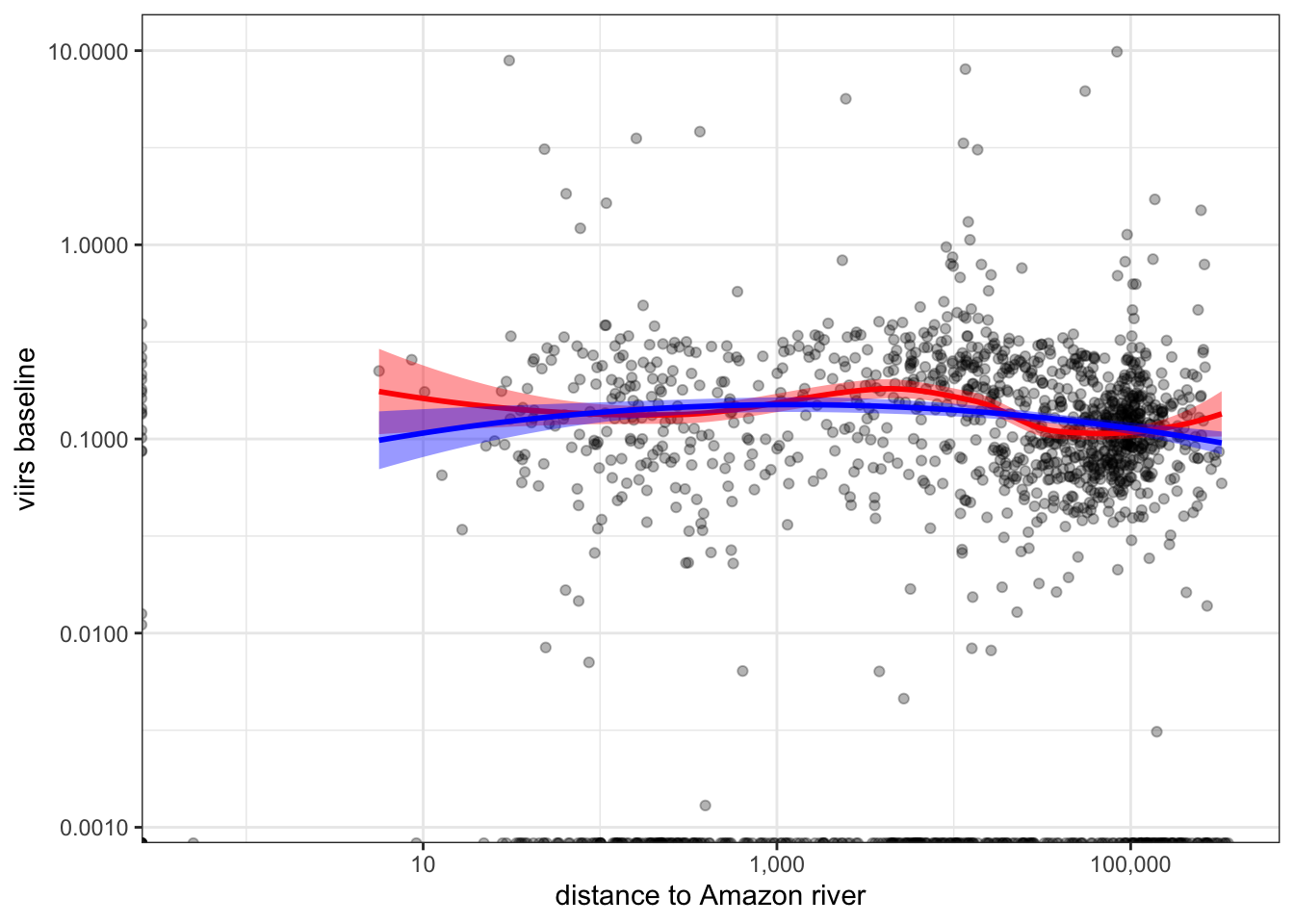
2.5.8 Popualtion vs. beta [Fig 3b]
f3b.p <- b %>%
ggplot(aes(x=nrohab, y=nl_viirs_2_max2)) +
geom_point(alpha=.3, shape = 3) +
scale_x_continuous(trans="log10", labels = scales::comma_format()) +
labs(x = "Population (hab.)", y = "VIIRS beta") +
#geom_smooth(method = "loess", col="red", fill="red") +
stat_smooth(method = "lm", formula = y ~ poly(x, 2), col="#FDE725FF", fill="#FDE725FF") +
theme_bw()
f3b <- ggMarginal(f3b.p, xparams = list(fill = "#481568FF", col = "#440154FF", alpha=.3),
yparams = list(fill = "#29AF7FFF", col = "#20A386FF", alpha=.3))
library(cowplot)
f3 <- plot_grid(f3a, f3b, labels = c("A","B"))
f3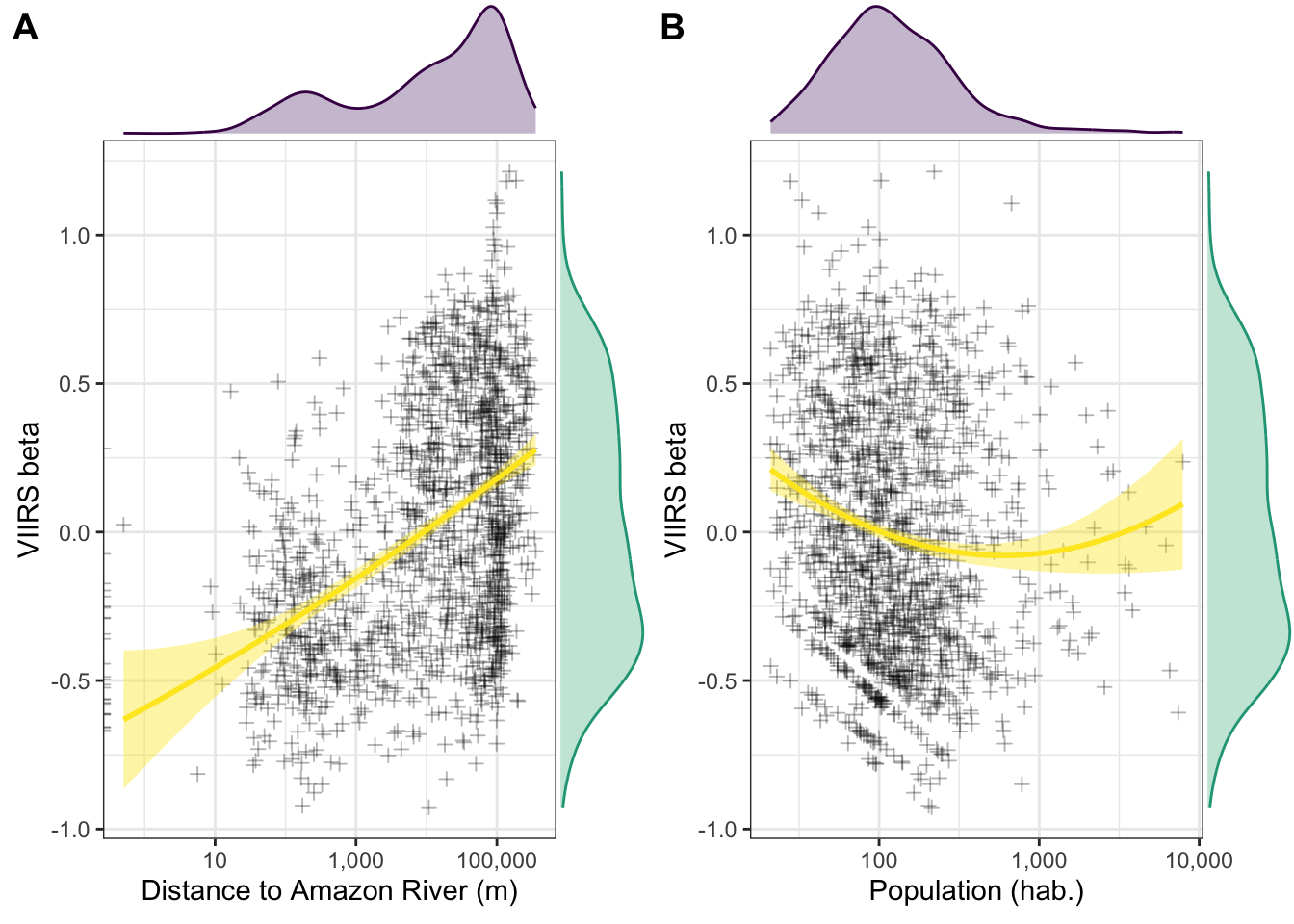
#ggsave2(f3, filename = "./_out/f3.png", width = 12, height = 7, dpi = 300)2.5.9 Popualtion vs. VIIRS baseline
b %>%
ggplot(aes(x=nrohab, y=nl_viirs_2)) +
geom_point(alpha=.3) +
scale_x_continuous(trans="log10", labels = scales::comma_format()) +
scale_y_continuous(trans="log10", labels = scales::comma_format()) +
labs(x = "Population", y = "viirs baseline") +
geom_smooth(method = "loess", col="red", fill="red") +
stat_smooth(method = "lm", formula = y ~ poly(x, 2), col="blue", fill="blue") +
theme_bw()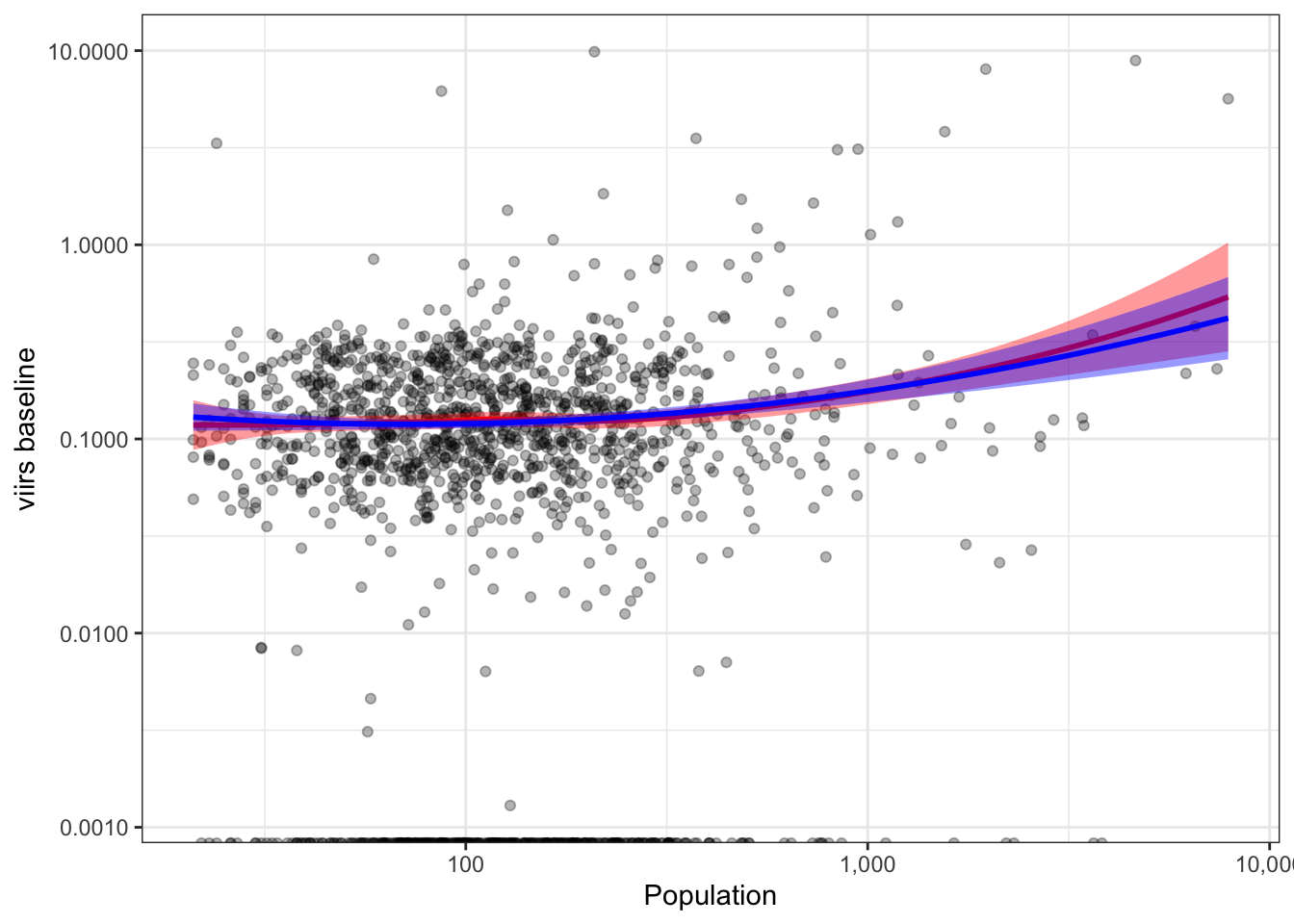
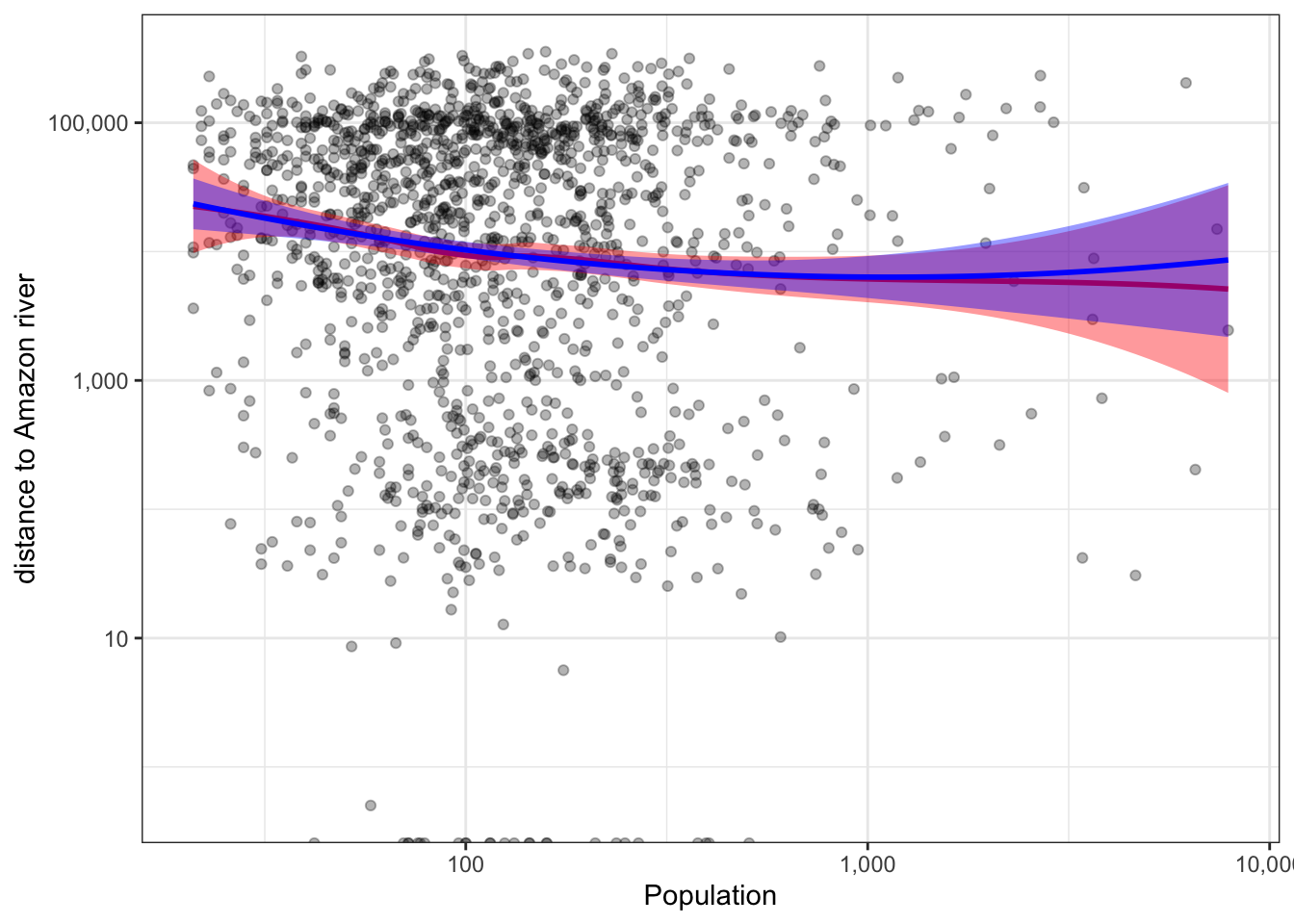
2.5.10 Popualtion vs. Distance to Amazon
b %>%
ggplot(aes(x=nrohab, y=distance)) +
geom_point(alpha=.3) +
scale_x_continuous(trans="log10", labels = scales::comma_format()) +
scale_y_continuous(trans="log10", labels = scales::comma_format()) +
labs(x = "Population", y = "distance to Amazon river") +
geom_smooth(method = "loess", col="red", fill="red") +
stat_smooth(method = "lm", formula = y ~ poly(x, 2), col="blue", fill="blue") +
theme_bw()