Chapter 4 Online Classes
4.1 Machine Learning
- Currently taking two courses on machine learning
- Practical Machine Learning with R on Coursera
- Applied Machine Learning via FAES
4.1.1 Practical Machine Learning
4.1.1.1 Why Preprocess?
Features can be very skewed
preProcessfunction in thecaretpackage allows for standardization/normalization of data- Can use a
preProcessobject to adjust other subsets (eg. test set) preProcessargument exists withintrainfunction
- Can use a
BoxCox transforms take continuous data, and attempts to make them look like normal data
Can also impute data
- Various methods such as
knnImpute
- Various methods such as
Training and test set must be processed in the same way
Test transformations will likely be imperfect
- Especially if the sets are collected at different times
Be careful when transforming factor variables
4.1.1.2 Covariate Creation
Two levels of covariate creation
- From raw data to covariate
- Converting text into token counts
- Transforming tidy covariates
Functional transformations (log, etc)
Level 1
- Depends on application
- summarization vs information loss
- when in doubt, stay on the side of more features
- can be automated, but use caution
Level 2
- More necessary for some methods (regression, svms)
- Only on the training set
- Discovery through EDA
- New covariates should be added to data frames (don’t removed original)
use
nearZeroVarfunction incaretto remove unimportant featureslibrary(splines)to create a bs basis- creation of polynomial variables to then fit
- fitting curves w/splines
4.1.1.3 Preprocessing with Principal Components Analysis
- Use with many correlated predictors
- Weighted combination of predictors
- Reduced predictors/noise
- Find a new set of variables that are uncorrelated and explain as much variance as possible
- Find the best matrix with fewer variables (lower rank) that explains the original data
- Related solutions - SVD and PCA
prcompin R- Can preprocess with PCA in caret
- Have to use same PCA in test set
- Make sure to transform first
4.1.1.4 Quiz 2
4.1.1.4.1 Question 1
4.1.1.4.2 Question 2
library(Hmisc)
library(tidyverse)
data(concrete)
set.seed(1000)
inTrain = createDataPartition(mixtures$CompressiveStrength, p = 3/4)[[1]]
training = mixtures[ inTrain,]
testing = mixtures[-inTrain,]Make a plot of the outcome (CompressiveStrength) versus the index of the samples. Color by each of the variables in the data set (you may find the cut2() function in the Hmisc package useful for turning continuous covariates into factors). What do you notice in these plots?
base_plot <- ggplot(concrete, aes(1:nrow(concrete), CompressiveStrength)) +
geom_point()
for (x in names(concrete)) {
print(
base_plot +
aes_string(col = x)
)
}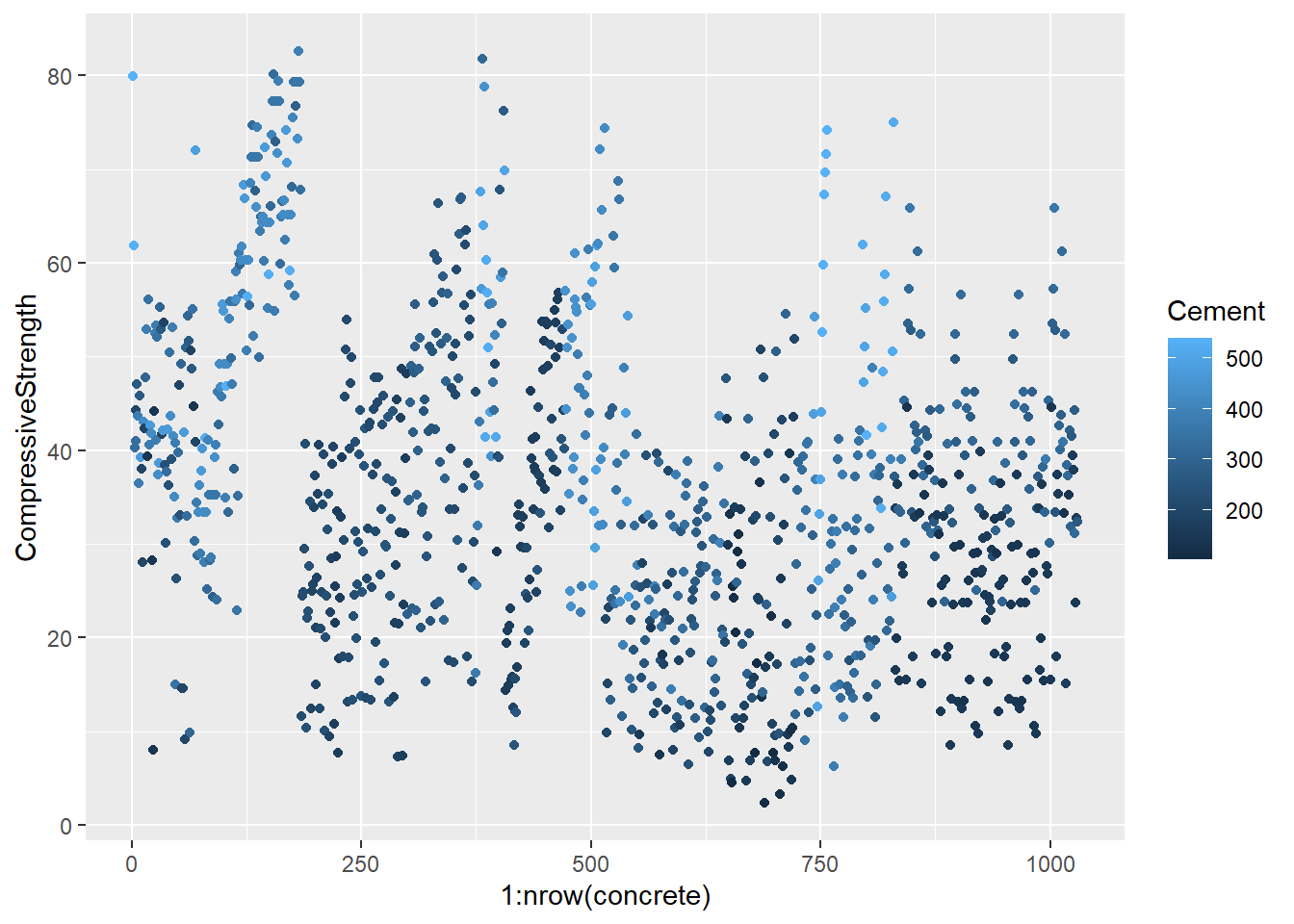
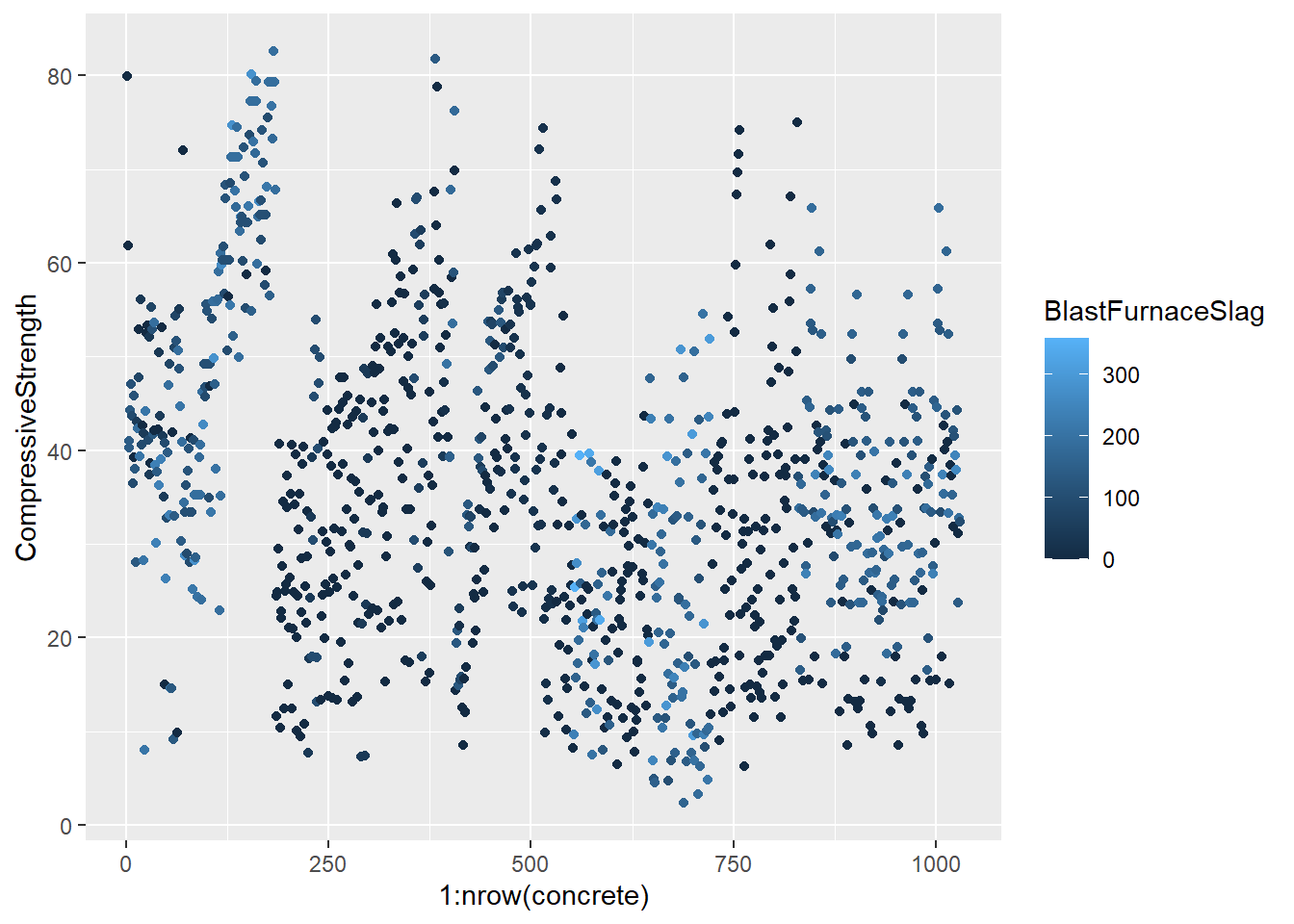
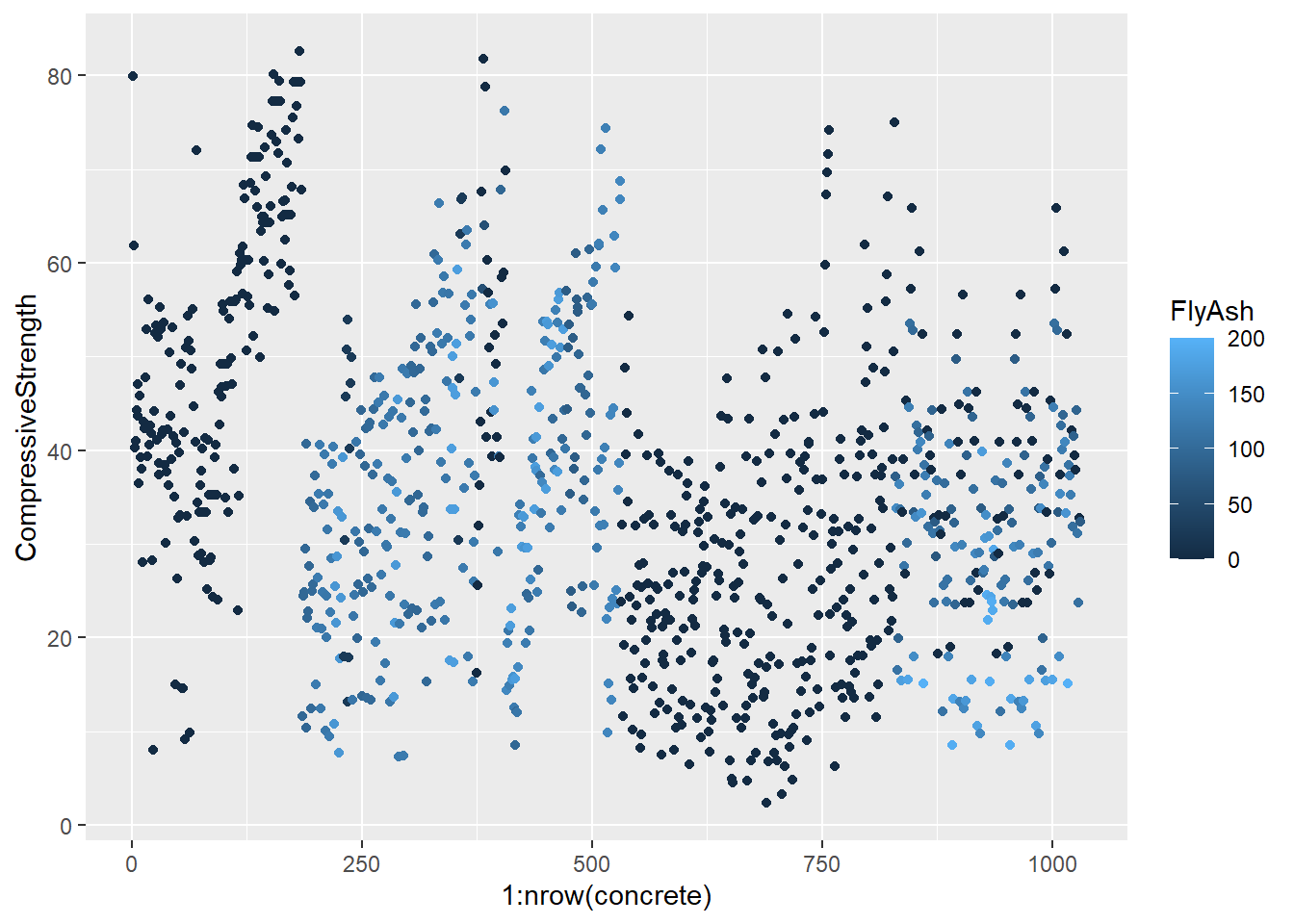
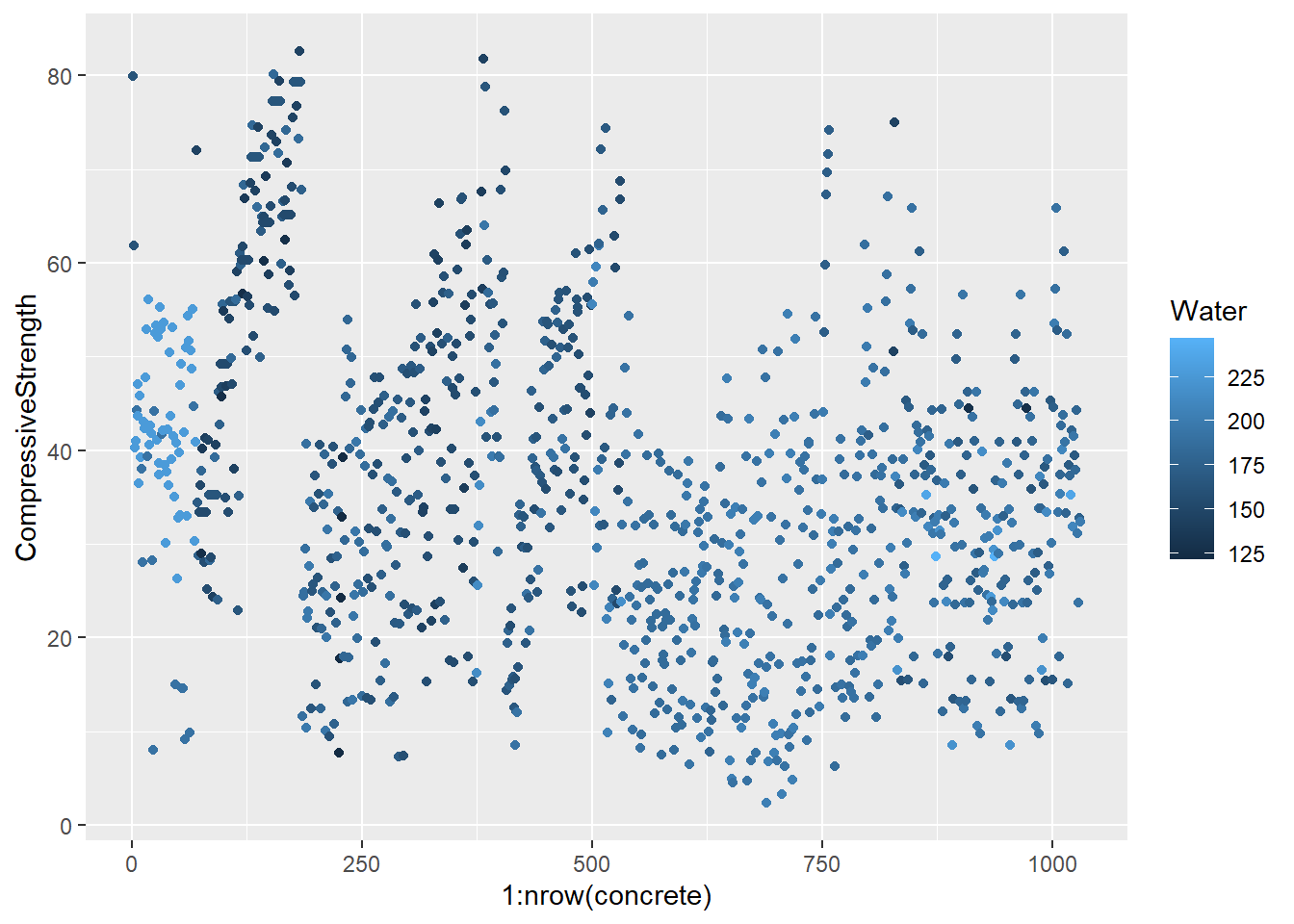
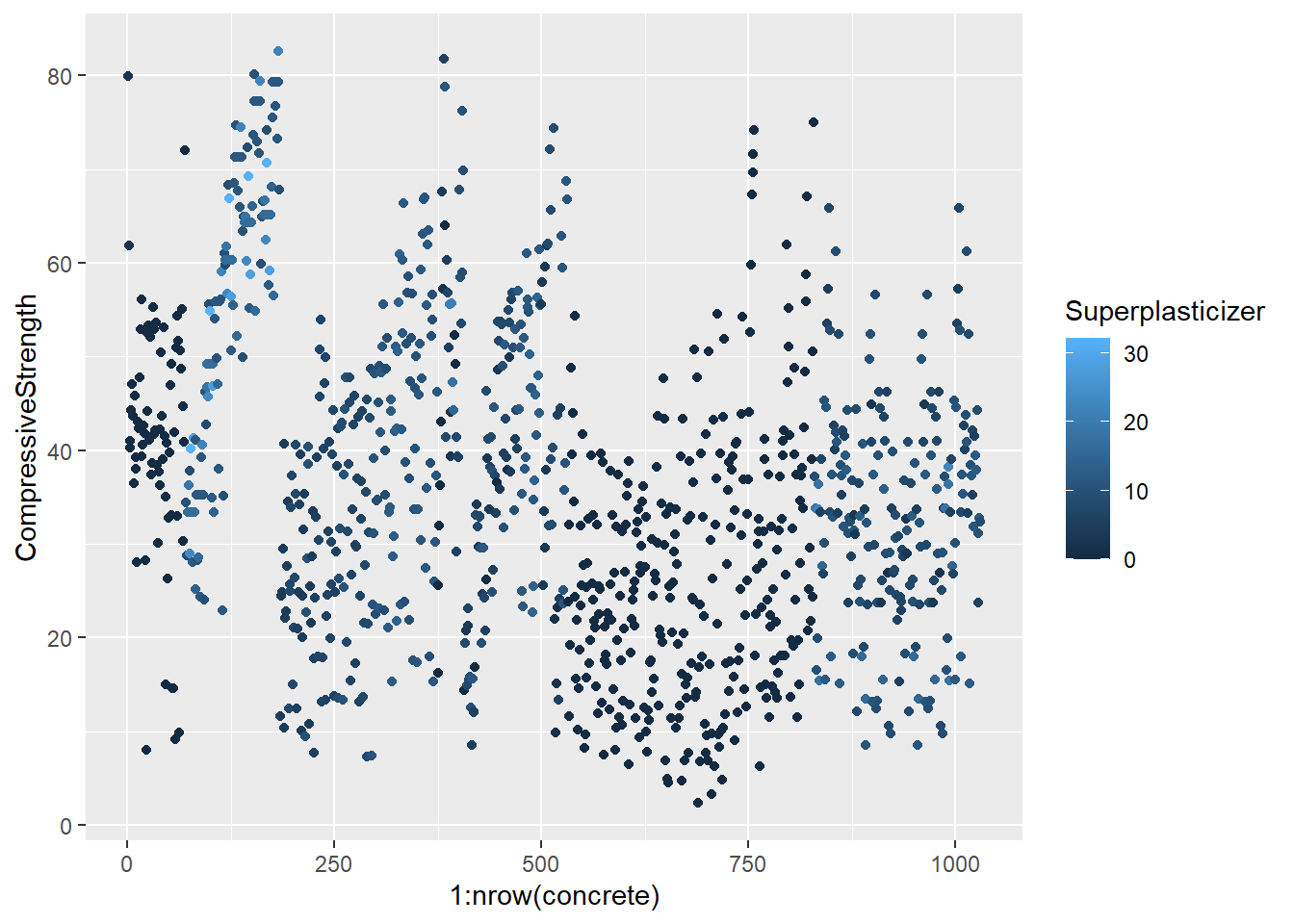
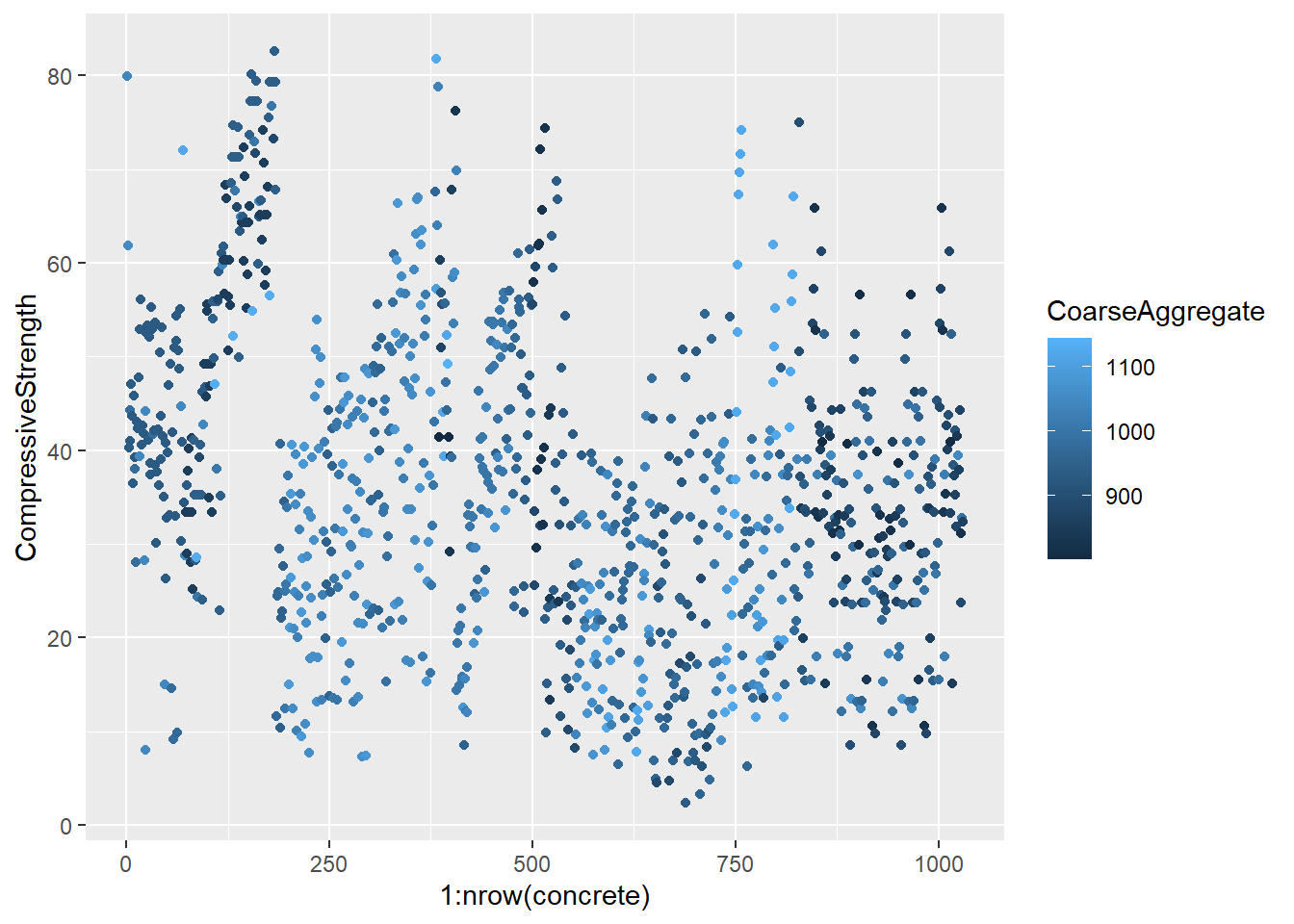
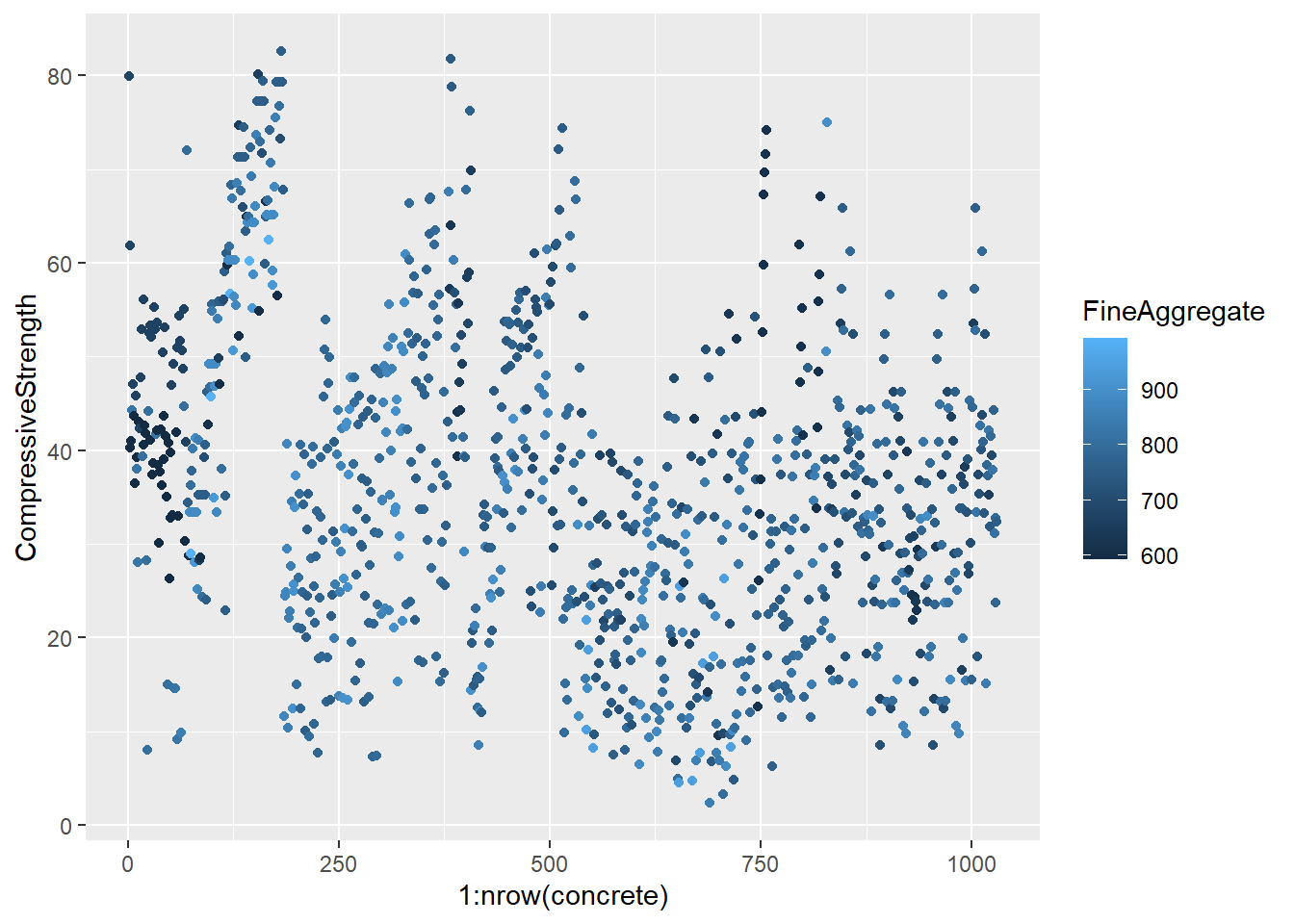
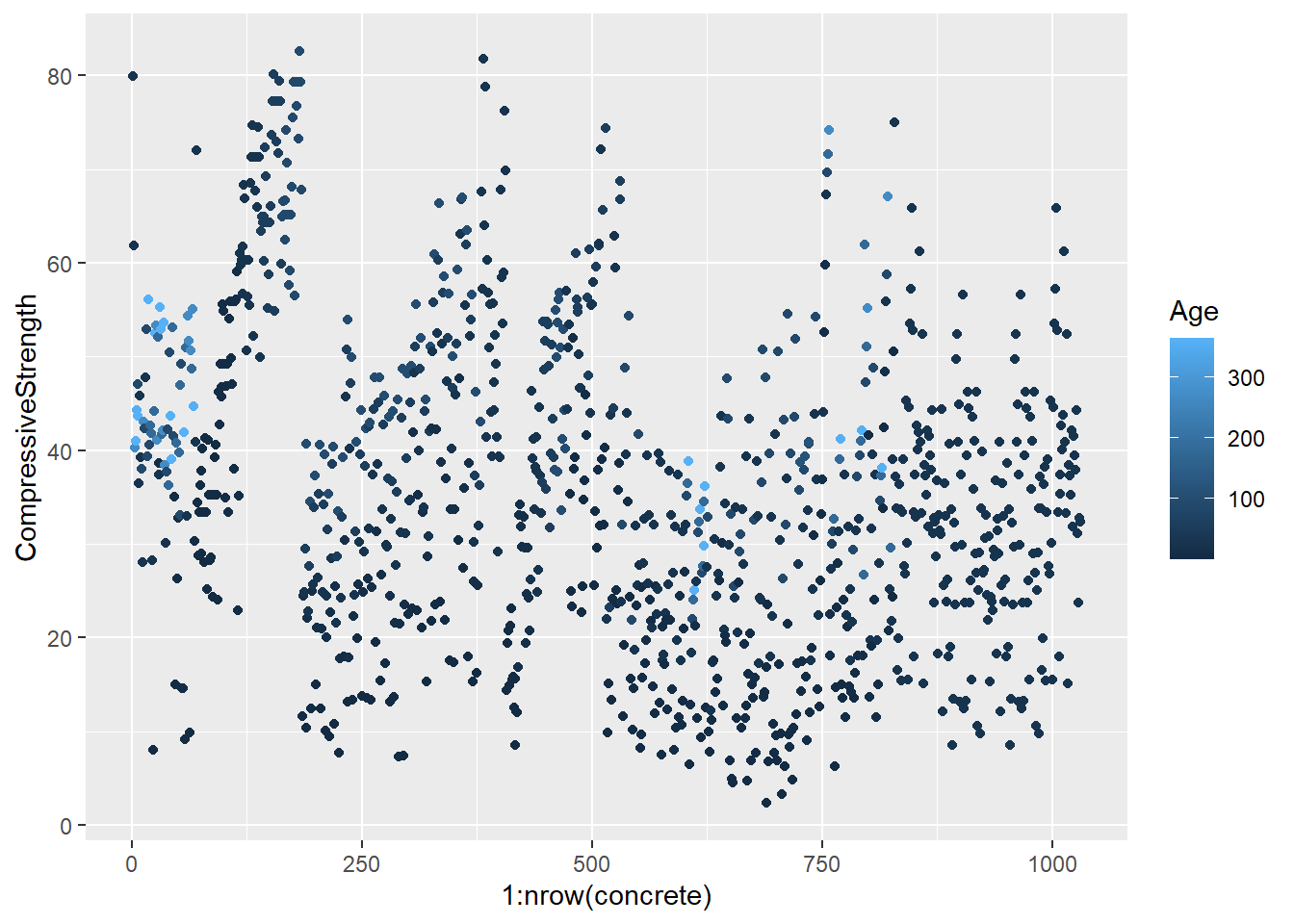
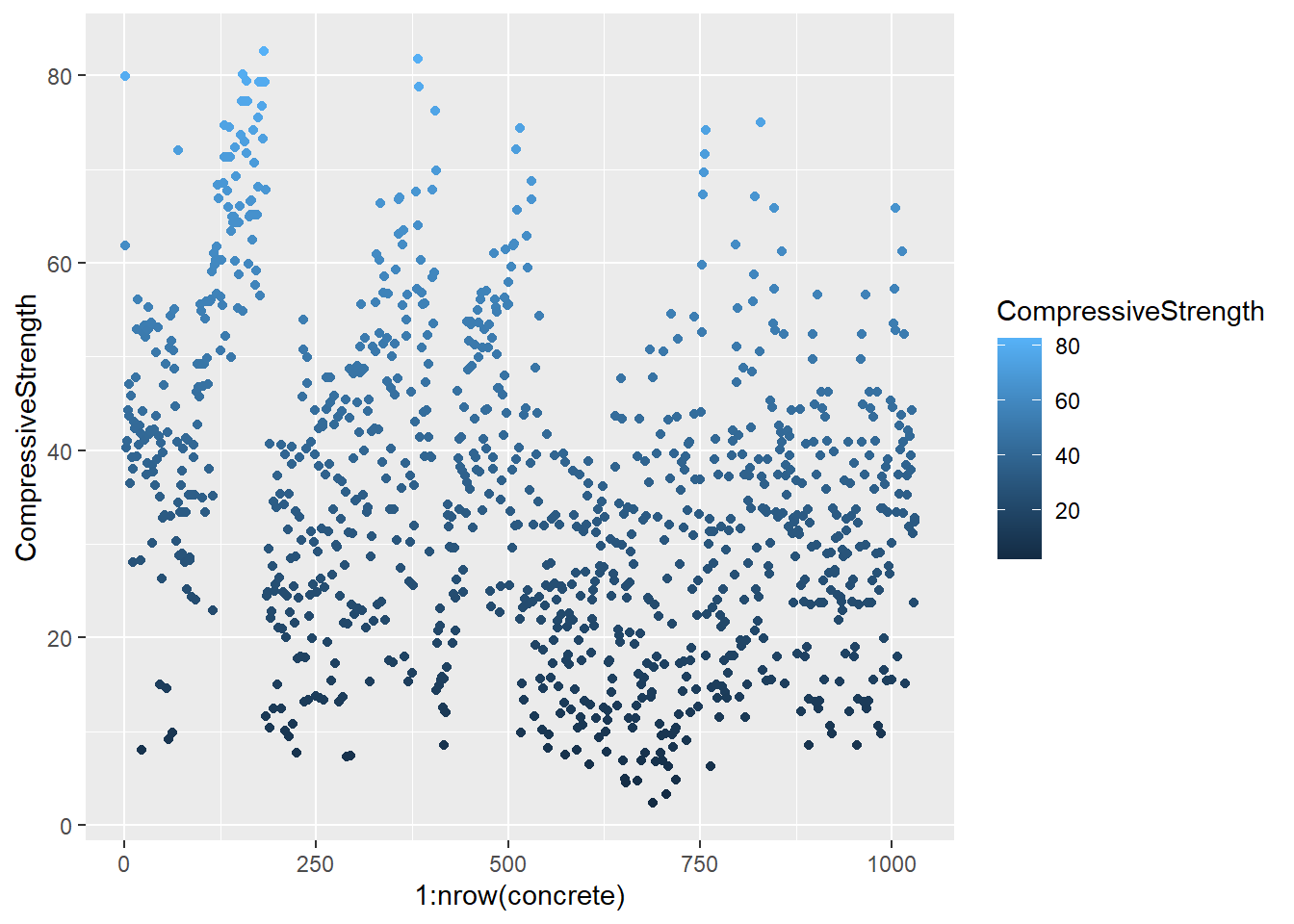
4.1.1.4.3 Question 3
data(concrete)
set.seed(1000)
inTrain = createDataPartition(mixtures$CompressiveStrength, p = 3/4)[[1]]
training = mixtures[ inTrain,]
testing = mixtures[-inTrain,]Make a histogram and confirm the SuperPlasticizer variable is skewed. Normally you might use the log transform to try to make the data more symmetric. Why would that be a poor choice for this variable?
## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.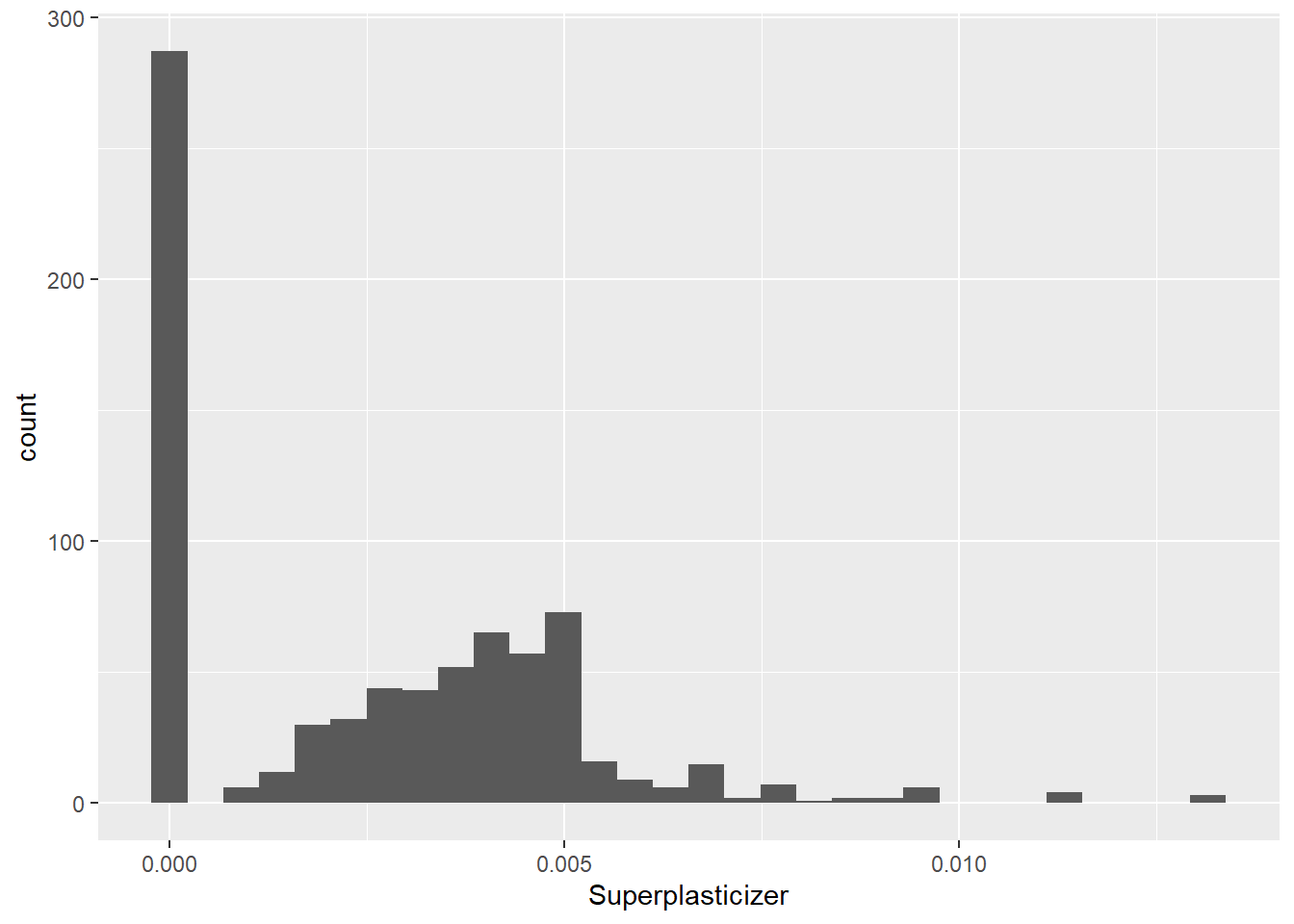
## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.## Warning: Removed 287 rows containing non-finite values (stat_bin).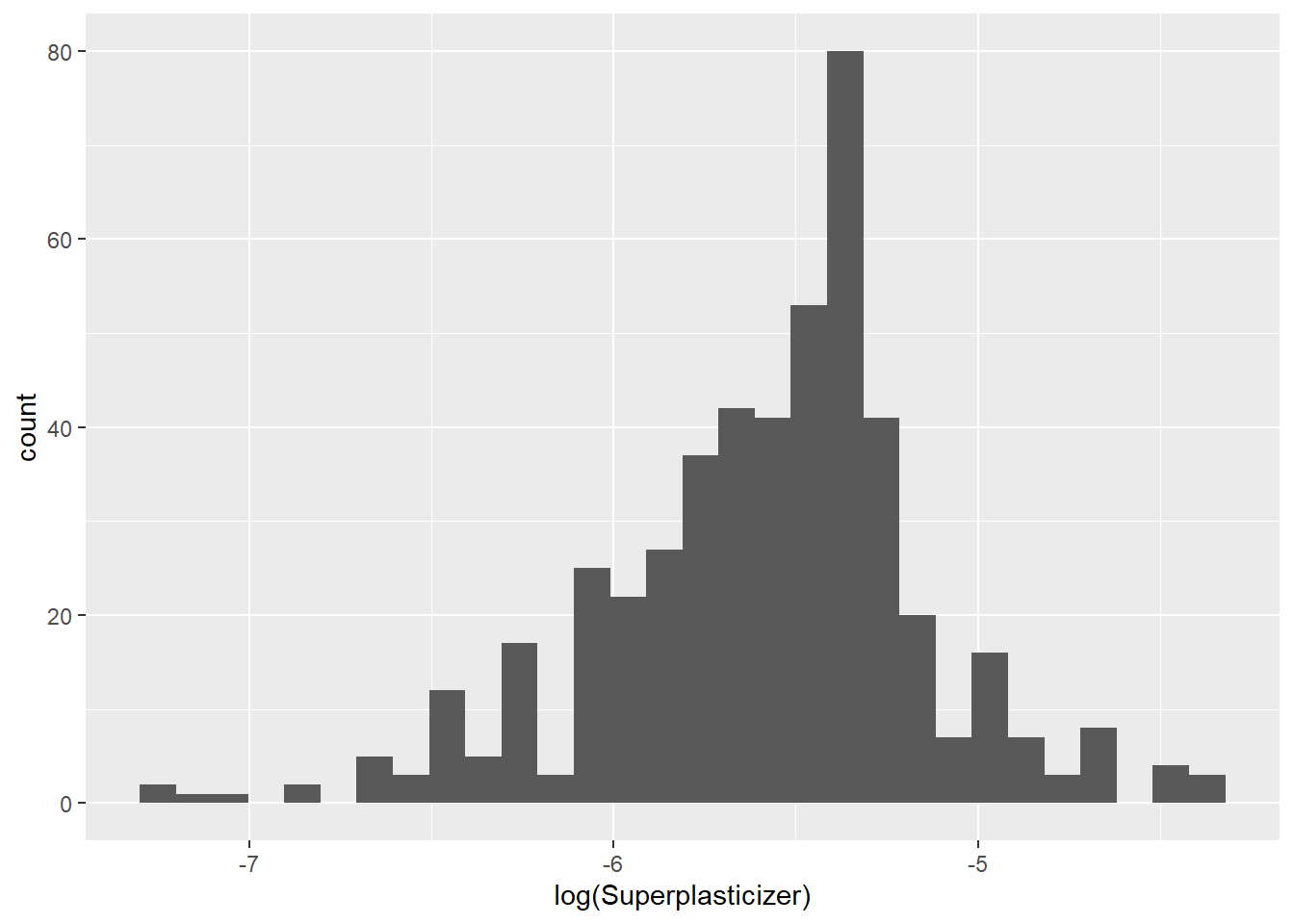
4.1.1.4.4 Problem 4
set.seed(3433)
data(AlzheimerDisease)
adData = data.frame(diagnosis,predictors)
inTrain = createDataPartition(adData$diagnosis, p = 3/4)[[1]]
training = adData[ inTrain,]
testing = adData[-inTrain,]Find all the predictor variables in the training set that begin with IL.
## [1] "IL_11" "IL_13" "IL_16" "IL_17E"
## [5] "IL_1alpha" "IL_3" "IL_4" "IL_5"
## [9] "IL_6" "IL_6_Receptor" "IL_7" "IL_8"Perform principal components on these variables with the preProcess() function from the caret package. Calculate the number of principal components needed to capture 80% of the variance. How many are there?
il_training <- training %>%
select(
starts_with("IL")
)
preProcess(il_training, method = "pca", thresh = 0.8)## Created from 251 samples and 12 variables
##
## Pre-processing:
## - centered (12)
## - ignored (0)
## - principal component signal extraction (12)
## - scaled (12)
##
## PCA needed 7 components to capture 80 percent of the variance