Chapter 8 Advanced options
8.1 Custom regional summaries
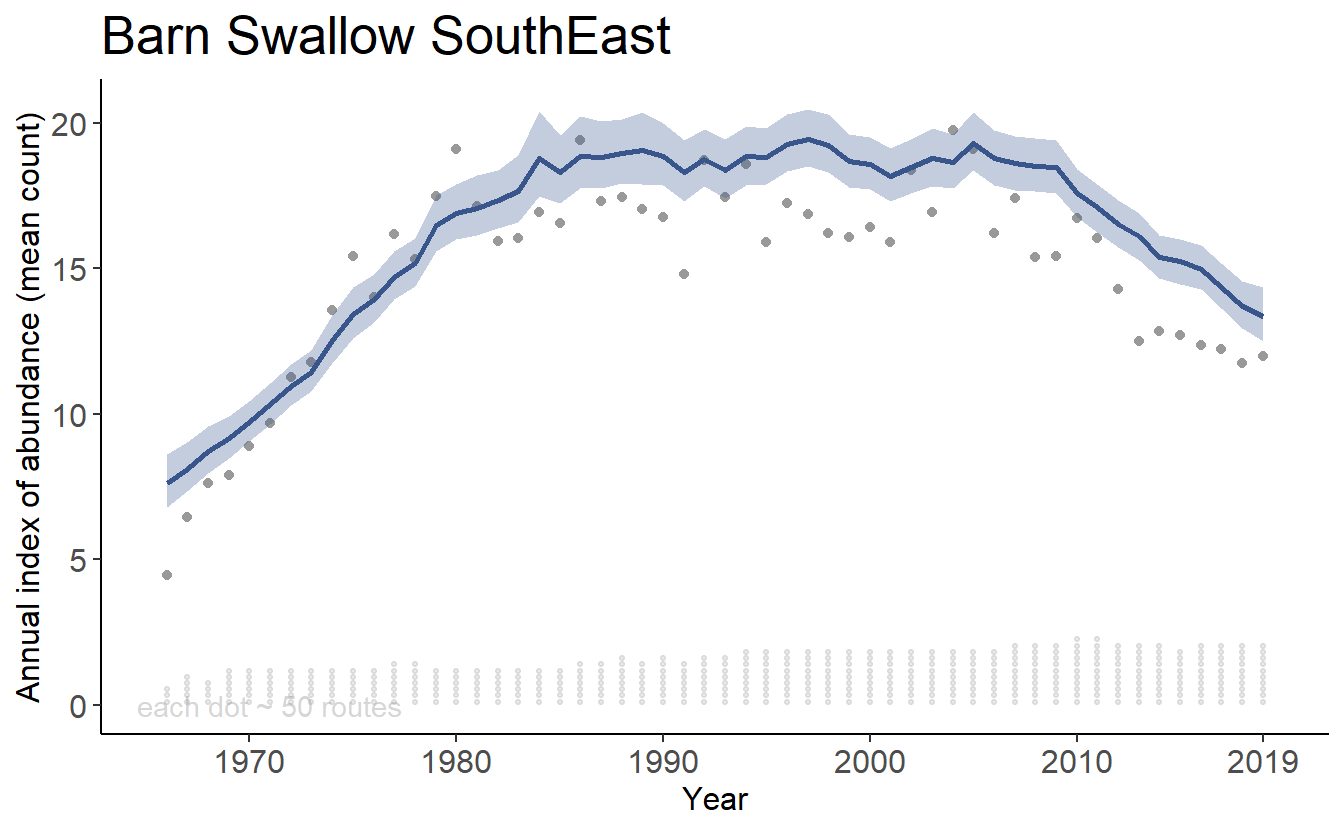
Yes, you can calculate the trend and trajectories for custom combinations of strata, such as a formal trend estimate for populations of Barn Swallow in the South East portion of their range (e.g., BCRs 21,24,25,26,27,35,36, and 37).
8.1.1 Define the custom regions as a collection of the existing strata.
First extract a dataframe that defines the original strata used in the analysis.
st_comp_regions <- get_composite_regions(strata_type = "bbs_cws")
knitr::kable(head(st_comp_regions))| prov_state | region | area_sq_km | national | bcr | Province_State | Country | bcr_by_country |
|---|---|---|---|---|---|---|---|
| AB | CA-AB-10 | 52565 | CA | 10 | Alberta | Canada | Canada-BCR_10 |
| AB | CA-AB-6 | 445136 | CA | 6 | Alberta | Canada | Canada-BCR_6 |
| AB | CA-AB-8 | 6987 | CA | 8 | Alberta | Canada | Canada-BCR_8 |
| AB | CA-AB-11 | 149352 | CA | 11 | Alberta | Canada | Canada-BCR_11 |
| AK | US-AK-1 | 9551 | US | 1 | ALASKA | United States of America | United States of America-BCR_1 |
| AK | US-AK-2 | 283405 | US | 2 | ALASKA | United States of America | United States of America-BCR_2 |
Then add a column to the dataframe that groups the original strata into the desired custom regions.
st_comp_regions$SouthEast <- ifelse(st_comp_regions$bcr %in% c(21,24:27,35:37),"SouthEast","Other")8.1.2 Use defined regions to estimate indices and trends
st_comp_regions can now be used as the dataframe input to the argument alt_region_names in generate_indices(), with “SouthEast” as the value for the argument regions. The relevant trends can be calculated using just the generate_trends() function.
library(patchwork)
custom_indices <- generate_indices(jags_mod = jags_mod,
jags_data = jags_data,
alt_region_names = st_comp_regions,
regions = "SouthEast")
tp = plot_indices(indices = custom_indices,
species = "Barn Swallow",
add_observed_means = TRUE,
add_number_routes = TRUE,
title_size = 20,
axis_title_size = 12,
axis_text_size = 12)print(tp[[1]])
print(tp[[2]])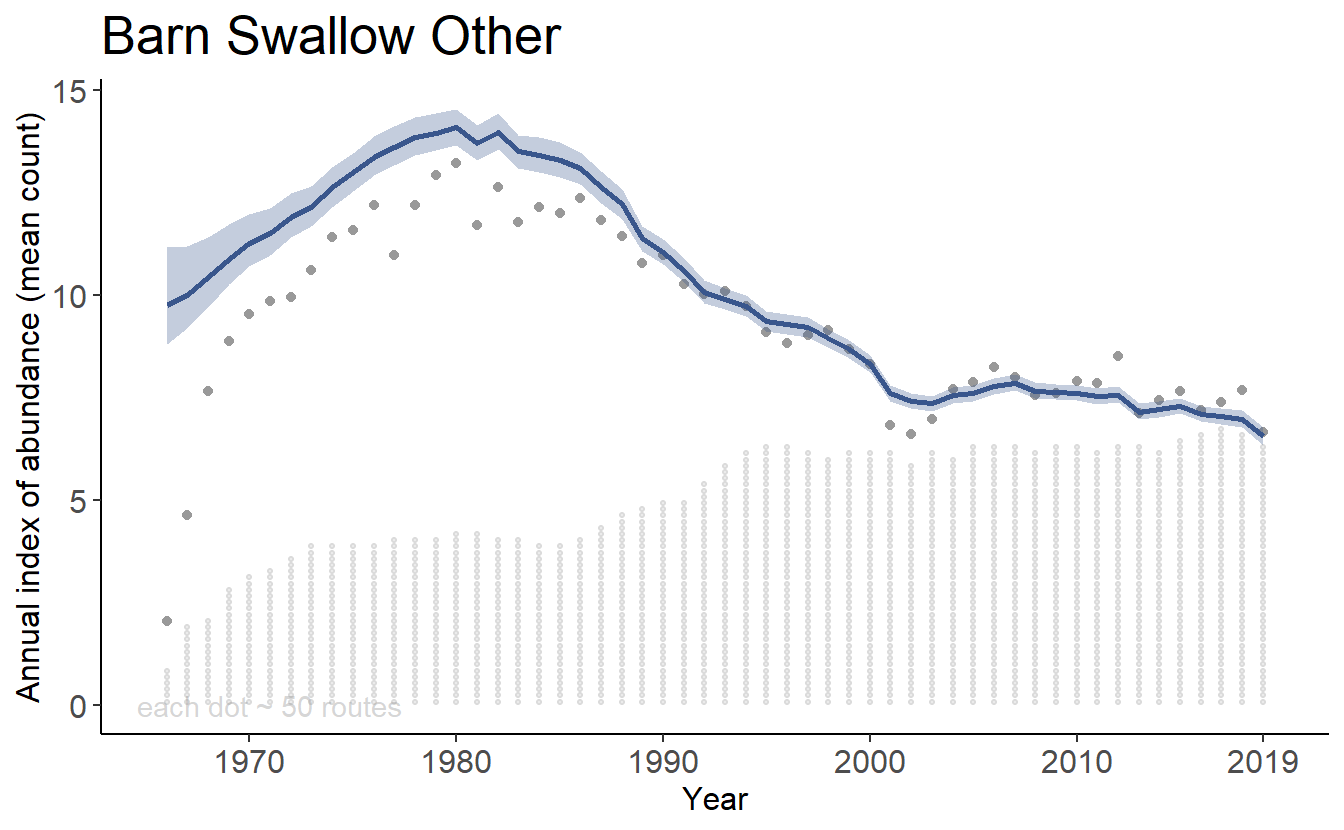
Calculate trends
custom_trends <- generate_trends(indices = custom_indices)
knitr::kable(custom_trends[,c(1,3,8,9,14)])8.2 Exporting the JAGS model
You can easily export any of the bbsBayes models to a text file.
model_to_file(model = "slope",
filename = "my_slope_model.txt")This is the best way to confirm all of the priors, and model structures.
8.3 Customizing the JAGS model and data
You can also modify the model text (e.g., try a different prior) and run the modified model
run_model <- function(... ,
model_file_path = "my_modified_slope_model.txt",
... )Or, you could add covariates. For example a GAM smooth to estimate the effect of the day of year on the observations, or an annual weather covariate, or… Then add the relevant covariate data to the jags_data object, and you’re off!
Here’s a GitHub repo with an example of a modified bbsBayes model. The modified model includes a series of GAM smooths to account for variation in counts over the course of the BBS observation season (mid-May through early July). In addition these seasonal smooths are allowed to vary by decade to account for possible shifts in phenology through time.
8.4 Comparing Models
Finally, bbsBayes can be used to run cross-validations. For example, the get_final_values() function is useful to provide an efficient starting point for a cross-validation runs, without having to wait for another full burn-in period.
Paper that includes an example of how to implement a cross-validation using bbsBayes.
Ornithological Applications: https://doi.org/10.1093/ornithapp/duaa065 Supplement: https://zenodo.org/badge/latestdoi/228419725
NOTE: although bbsBayes includes functions to calculate WAIC, recent work has shown that WAIC performs very poorly with the BBS data (https://doi.org/10.1650/CONDOR-17-1.1). We recommend a k-fold cross-validation approach, as in the above zenodo archive.