Chapter 3 Applications
3.1 Application I: Individual-level data
The dataset used here is a subset of data taken from ‘MetaboAnalyst’ (Xia et al. 2009) example datasets in the statistical meta-analysis section. These data are from a study investigating biomarkers of adenocarcinoma in blood and this dataset consists of a total of 14 metabolites (N = 14) and 172 samples collected from two studies (K = 2). We will use `MetaHD’ for combining summary estimates obtained from these multiple metabolomic studies.
This real dataset is available in the `MetaHD’ package and can be loaded using the following command.
As explained in Section 3.1 `MetaHDInput’ function can then be used to calculate the observed effect sizes and their associated variances and within-study covariance matrices as shown below.
## met1 met2 met3 met4 met5
## study1 -0.08509765 0.0693374 0.21205246 0.1555140 -0.07131115
## study2 -0.19103975 0.1985993 0.05266387 0.3042082 -0.08507508
## met6 met7 met8 met9 met10
## study1 -0.010729262 -0.06351313 0.07239534 -0.02854481 -0.1874413
## study2 -0.002200277 -0.02427915 -0.08497286 -0.10315078 -0.1196099
## met11 met12 met13 met14
## study1 0.07670815 -0.04170964 0.3378766 -0.20944830
## study2 0.07105905 0.21213759 0.2136121 -0.03345043## met1 met2 met3 met4
## met1 0.0194438538 0.0005017922 -0.0025219163 -0.002766591
## met2 0.0005017922 0.0161291933 0.0042058671 0.019843118
## met3 -0.0025219163 0.0042058671 0.0078052190 0.007030307
## met4 -0.0027665912 0.0198431182 0.0070303073 0.074406890
## met5 0.0049925375 0.0025359133 -0.0012859676 -0.003247293
## met6 0.0036952818 0.0035899796 0.0007582538 0.004244171
## met5 met6 met7 met8 met9
## met1 0.004992537 0.0036952818 0.0036649961 0.0014738763 0.002466527
## met2 0.002535913 0.0035899796 0.0023004763 0.0009291760 0.003908964
## met3 -0.001285968 0.0007582538 -0.0002967142 0.0010746462 0.001839553
## met4 -0.003247293 0.0042441714 0.0033677211 0.0053704235 0.004464203
## met5 0.007725915 0.0031553517 0.0026034856 -0.0006582206 0.002925312
## met6 0.003155352 0.0035383608 0.0025416742 0.0003542152 0.003045991
## met10 met11 met12 met13 met14
## met1 0.006057970 0.003742686 0.0058389366 -0.017656900 0.006189749
## met2 0.001622693 0.006128897 0.0057661483 -0.015584915 0.002615291
## met3 -0.002939468 0.002551313 0.0003375944 0.001663327 -0.001850528
## met4 -0.001121570 0.005545238 0.0033374745 -0.012707585 0.001019416
## met5 0.005845677 0.004305489 0.0060072612 -0.020351263 0.004988612
## met6 0.003098343 0.003080440 0.0039539767 -0.012411644 0.003099160MetaHD model can now be fitted as follows:
# Multivariate high dimensional meta analyis.
model1 <- MetaHD(Y, Slist, method = "reml", bscov = "unstructured")
model1$estimate## [1] -0.1294875230 0.1390541472 0.1187040998 0.2031757519
## [5] -0.0406585464 -0.0001290338 -0.0338842144 -0.0052315949
## [9] -0.0474366927 -0.1240326912 0.0946724585 0.1105743431
## [13] 0.1909300556 -0.1084060240## met1 met2 met3 met4 met5 met6
## 0.10014867 0.08615324 0.09690759 0.25810338 0.36261181 0.99461969
## met7 met8 met9 met10 met11 met12
## 0.28079034 0.92795765 0.41512986 0.01378248 0.03066823 0.35119400
## met13 met14
## 0.01358336 0.12513848When both within and between study correlations are set to zero, MetaHD reduces to the usual random effects model analysis of individual metabolites (i.e. univariate meta-analyses). Additionally, when the between-study variances are also set to zero, then MetaHD reduces to a fixed-effects model univariate meta-analysis. We can perform univariate random-effects and univariate fixed-effects meta-analysis in MetaHD as follows:
diag_Slist <- list()
K <- nrow(Y)
for (k in 1:K) {
diag_Slist[[k]] <- diag(diag(Slist[[k]]))
}
# Univariate random-effects meta-analysis
model2 <- MetaHD(Y, diag_Slist, method = "reml", bscov = "diag")
model2$estimate## [1] -0.152811460 0.137084798 0.125385177 0.236479720 -0.079156886
## [6] -0.003401107 -0.041615382 -0.005821595 -0.067093064 -0.145747152
## [11] 0.073429624 0.097638044 0.224771586 -0.115494377## [1] 0.068106515 0.117949995 0.112626720 0.199119540 0.169892097
## [6] 0.878896872 0.281697786 0.940930454 0.297320202 0.012461083
## [11] 0.161918116 0.439516841 0.009507967 0.190310323# Univariate fixed-effects meta-analysis
model3 <- MetaHD(Y, diag_Slist, method = "fixed")
model3$estimate## [1] -0.152812327 0.137090057 0.119263841 0.236485102 -0.079158213
## [6] -0.003400281 -0.041614290 -0.004927685 -0.067093550 -0.145743651
## [11] 0.073429398 0.139309196 0.224769883 -0.102735148## [1] 0.068095484 0.117624861 0.036764299 0.198923825 0.169579632
## [6] 0.878875296 0.281592165 0.914730005 0.297222122 0.012441239
## [11] 0.161809425 0.011347289 0.009502382 0.034970100When missing data are available, we can use MetaHD to handle missing values by setting the argument “impute.na = TRUE”. This replaces the missing effect sizes and variances by zero and a large constant value respectively, so that the missing outcome is allocated a lower weight in the meta-analysis.
3.2 Application II: Summary-level data
In this example, we use a simulated dataset as described by (Liyanage et al. 2024). Here we have prepared separate data frames for observed effect sizes and within-study covariance matrices in the ‘simdata.1’ dataset as follows..
## metabolite_1 metabolite_2 metabolite_3 metabolite_4
## study_1 0.231755235 -0.9377271 -3.306843 0.28142576
## study_2 0.305408170 -1.2421302 -3.547565 0.66254064
## study_3 -0.006523691 -1.6762418 -3.631345 0.68023162
## study_4 0.283789796 -1.2533024 -3.545591 0.08533545
## study_5 -0.748073950 -2.5502749 -4.386687 -1.08561122
## study_6 -0.805069877 -2.2028554 -3.684388 -0.42021699
## metabolite_5 metabolite_6 metabolite_7 metabolite_8
## study_1 -0.5547623 2.822603 -1.268180 2.593808
## study_2 -0.3162607 2.660019 -1.434220 2.558514
## study_3 -0.1846545 2.421983 -1.085327 3.099015
## study_4 -1.0310925 2.245691 -1.448434 2.231622
## study_5 -1.7532039 1.245282 -2.156295 1.503325
## study_6 -1.3547757 2.730196 -1.792440 1.812767
## metabolite_9 metabolite_10 metabolite_11 metabolite_12
## study_1 -0.3936202 -0.6887844 1.25723073 -0.1669118
## study_2 -0.2623882 -0.0245477 1.50515960 -0.1288290
## study_3 -0.2417435 0.3493618 1.02398696 -0.4566538
## study_4 -0.6266361 -0.4590747 1.01206691 -0.4866803
## study_5 -1.4695383 -1.3845496 0.05619328 -1.0199653
## study_6 -0.4075956 -0.9052131 0.11930101 -0.9310851
## metabolite_13 metabolite_14 metabolite_15 metabolite_16
## study_1 1.4819259 2.067361 -0.3992064 2.035879
## study_2 1.6942841 1.696545 -0.5495592 1.757267
## study_3 1.2831249 2.635289 -0.6367507 2.208511
## study_4 1.2184663 2.340882 -0.8044879 1.971764
## study_5 0.6820576 0.597100 -1.3620158 1.162320
## study_6 1.1849203 1.426632 -1.2488842 1.383772
## metabolite_17 metabolite_18 metabolite_19 metabolite_20
## study_1 1.6594976 1.4785488 2.025255 1.6325979
## study_2 1.5286110 1.4712913 2.068347 1.7831811
## study_3 1.2437205 1.7993070 1.877336 1.0564191
## study_4 0.6971900 1.3849356 2.472191 1.4691468
## study_5 0.5003652 0.3460756 1.408792 0.6553110
## study_6 1.3982598 0.8289669 1.587887 0.7490209
## metabolite_21 metabolite_22 metabolite_23 metabolite_24
## study_1 -0.31458426 0.24703204 3.177385 1.5929012
## study_2 -0.59096083 -0.18343696 2.512141 1.2888813
## study_3 -0.24805374 -0.08827114 3.071959 0.7913664
## study_4 -0.08455623 -0.18155226 2.534846 1.0723134
## study_5 -1.20477797 -1.12187511 1.513707 0.2390821
## study_6 -0.84545412 -0.58702267 2.308492 0.3502428
## metabolite_25 metabolite_26 metabolite_27 metabolite_28
## study_1 -0.005622678 -2.648980 -0.8436625 -0.904995
## study_2 0.139087964 1.797236 -0.4594717 -1.428674
## study_3 0.724998374 -3.313190 -1.0387204 -1.281120
## study_4 -0.258109359 -3.528483 -0.8659648 -1.185520
## study_5 -1.276384331 -4.118482 -2.0616843 -2.203919
## study_6 -0.838241155 -3.853180 -0.9640716 -1.899273
## metabolite_29 metabolite_30
## study_1 3.653576 5.543804
## study_2 3.077779 5.607509
## study_3 3.045154 5.590184
## study_4 4.045979 5.222089
## study_5 2.531608 5.210625
## study_6 3.069246 5.242326## metabolite_1 metabolite_2 metabolite_3 metabolite_4
## metabolite_1 0.2051971 0.2360390 0.1451389 0.2538377
## metabolite_2 0.2360390 0.3914241 0.2185096 0.2999849
## metabolite_3 0.1451389 0.2185096 0.1429655 0.2067593
## metabolite_4 0.2538377 0.2999849 0.2067593 0.3705385
## metabolite_5 0.2159263 0.2919311 0.1722842 0.2909224
## metabolite_6 0.3196974 0.3981609 0.2568697 0.4433588
## metabolite_5 metabolite_6 metabolite_7 metabolite_8
## metabolite_1 0.2159263 0.3196974 0.1667637 0.2076705
## metabolite_2 0.2919311 0.3981609 0.2405715 0.2527721
## metabolite_3 0.1722842 0.2568697 0.1591633 0.1504863
## metabolite_4 0.2909224 0.4433588 0.2554422 0.2650113
## metabolite_5 0.3120427 0.3949941 0.2104929 0.2520871
## metabolite_6 0.3949941 0.5994060 0.3069871 0.3358156
## metabolite_9 metabolite_10 metabolite_11 metabolite_12
## metabolite_1 0.08103280 0.11133998 0.1579164 0.2122468
## metabolite_2 0.12311977 0.15263329 0.2037668 0.2946624
## metabolite_3 0.07664409 0.09779216 0.1147814 0.1891951
## metabolite_4 0.11012275 0.15222817 0.1918580 0.2831199
## metabolite_5 0.09973932 0.14457419 0.1842593 0.2537483
## metabolite_6 0.13793029 0.19313481 0.2576985 0.3589582
## metabolite_13 metabolite_14 metabolite_15 metabolite_16
## metabolite_1 0.1870629 0.2068235 0.2090655 0.1991912
## metabolite_2 0.2341660 0.3079808 0.2931316 0.2709879
## metabolite_3 0.1460312 0.1852916 0.1698101 0.1665578
## metabolite_4 0.2571253 0.2892108 0.2479776 0.2599880
## metabolite_5 0.2475945 0.2650591 0.2702558 0.2520948
## metabolite_6 0.3438236 0.3801845 0.3523785 0.3548082
## metabolite_17 metabolite_18 metabolite_19 metabolite_20
## metabolite_1 0.11785788 0.2532525 0.11496091 0.1299937
## metabolite_2 0.16230953 0.2834488 0.14725054 0.1725120
## metabolite_3 0.09688847 0.1828497 0.09486947 0.1084088
## metabolite_4 0.15284705 0.3259542 0.15936720 0.1823390
## metabolite_5 0.13001982 0.3052326 0.13993093 0.1637614
## metabolite_6 0.19915187 0.4357530 0.20217217 0.2420832
## metabolite_21 metabolite_22 metabolite_23 metabolite_24
## metabolite_1 0.1487927 0.2316234 0.2108364 0.2635543
## metabolite_2 0.2002373 0.3218278 0.3002607 0.3586782
## metabolite_3 0.1201728 0.1890557 0.1783501 0.2206520
## metabolite_4 0.1959123 0.2944990 0.2774928 0.3359515
## metabolite_5 0.1944129 0.2788441 0.2557165 0.3522710
## metabolite_6 0.2604766 0.4135079 0.3512251 0.4720034
## metabolite_25 metabolite_26 metabolite_27 metabolite_28
## metabolite_1 0.2442990 0.2897116 0.1090710 0.2335329
## metabolite_2 0.3305478 0.3275086 0.1323510 0.2954209
## metabolite_3 0.1977199 0.2085774 0.0808657 0.1835814
## metabolite_4 0.2996849 0.3678765 0.1407640 0.2946390
## metabolite_5 0.2888747 0.3518957 0.1330913 0.2770655
## metabolite_6 0.3845858 0.4956480 0.1689191 0.3889943
## metabolite_29 metabolite_30
## metabolite_1 0.1896202 0.1358149
## metabolite_2 0.2487014 0.2008984
## metabolite_3 0.1600222 0.1192158
## metabolite_4 0.2564001 0.1816158
## metabolite_5 0.2196668 0.1739497
## metabolite_6 0.3132906 0.2299339Now, we can carry out the meta-analysis using MetaHD as follows.
## Multivariate random-effects meta analyis
model_MetaHD <- MetaHD(Y=Observed_effects,
Slist=Slist,
method = "reml",
bscov = "unstructured")
print(model_MetaHD$estimate) ## [1] -0.1661335 -1.9809593 -3.7329305 -0.3676187 -1.2805349 2.2047143
## [7] -1.8351657 1.9742259 -0.7673506 -0.9405747 0.6333586 -0.8693423
## [13] 1.0744949 1.3249918 -1.2437328 1.4046179 1.1997847 0.8963671
## [19] 1.7543779 1.1155858 -0.8517850 -0.6183473 2.4147259 0.5123058
## [25] -0.8787377 -3.6860782 -1.2630573 -1.7269780 3.0223912 5.5121450# TRUE VALUES ARE:
true_values<-c(-0.1675687 , -1.9658875 ,-3.7501346, -0.3722893, -1.2669714, 2.1815949, -1.8274545, 2.0032794, -0.7985332 ,-0.9362461 , 0.6539242 ,-0.8254938, 1.1240729, 1.3267165, -1.2057946,1.3967554, 1.1916929 , 0.9041837, 1.7934879, 1.1444330, -0.8233060, -0.5886543, 2.4371479, 0.4882229, -0.8903039, -3.6956073, -1.2576506, -1.7221614, 3.0298406, 5.4704779)
print(true_values)## [1] -0.1675687 -1.9658875 -3.7501346 -0.3722893 -1.2669714 2.1815949
## [7] -1.8274545 2.0032794 -0.7985332 -0.9362461 0.6539242 -0.8254938
## [13] 1.1240729 1.3267165 -1.2057946 1.3967554 1.1916929 0.9041837
## [19] 1.7934879 1.1444330 -0.8233060 -0.5886543 2.4371479 0.4882229
## [25] -0.8903039 -3.6956073 -1.2576506 -1.7221614 3.0298406 5.4704779In cases where the within-study correlations are not available, these could be estimated from the data as follows:
# If within-study correlations are not available, use only the variances and enter these values in a K x N matrix
Smat <- matrix(0, nrow = K, ncol = N)
for (i in 1:K) {
Smat[i, ] <- diag(Slist[[i]])
}
model_MetaHD_est.wscor<- MetaHD(Observed_effects ,
Smat,
method = "reml",
est.wscor = TRUE)Univariate random-effects meta analysis can be carried out as follows:
#Create matrices containing variances
diag_Slist <- list()
for (k in 1:K) {
diag_Slist[[k]] <- diag(diag(Slist[[k]]))
}
# Univariate random-effects meta-analysis
model_random <- MetaHD(Observed_effects ,
diag_Slist,
method = "reml",
bscov = "diag")Univariate fixed-effects meta analysis can be carried out as follows:
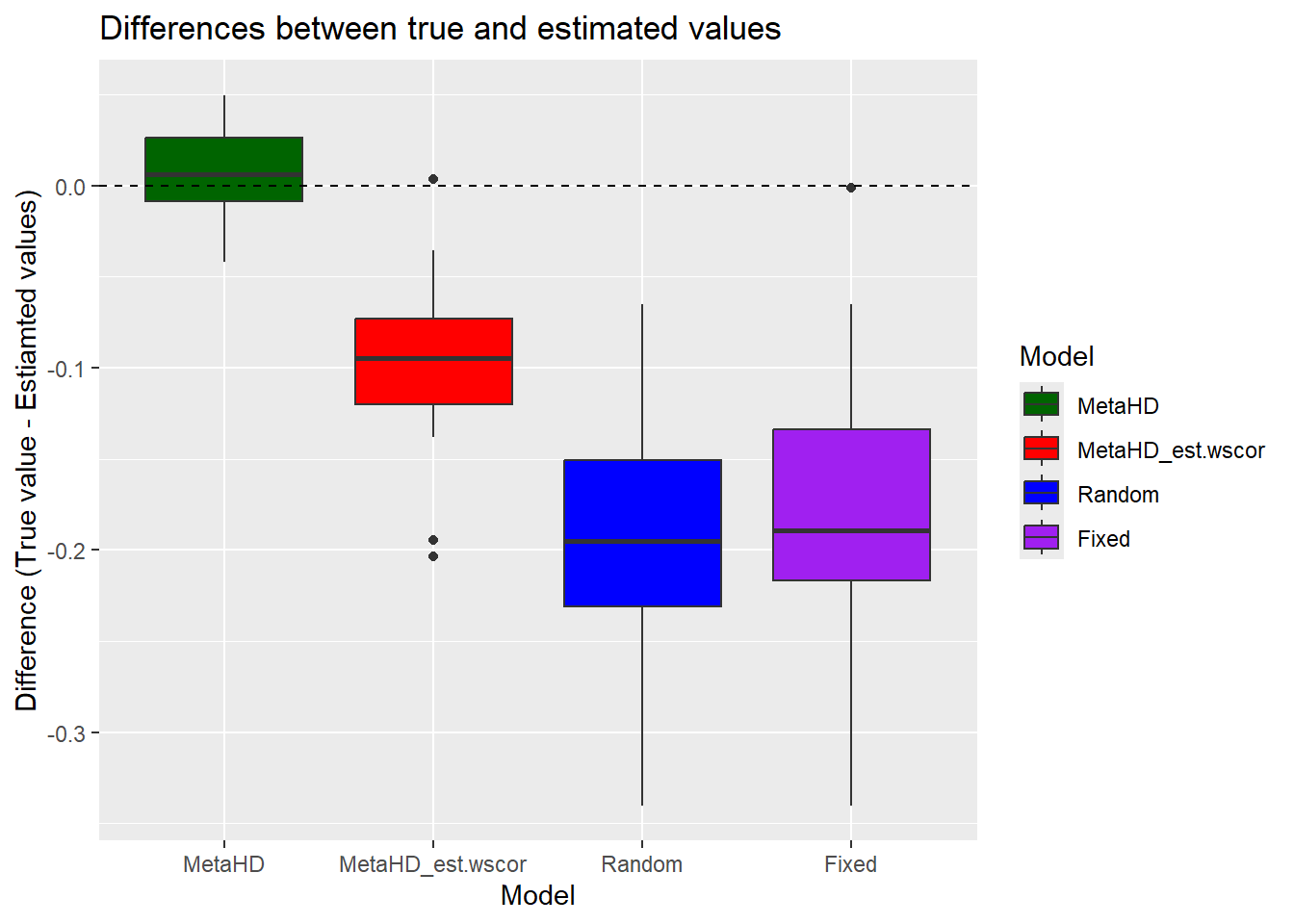
To compare the estimated values with the true values:
# Create a data frame of the differences
df <- data.frame(
Model = factor(rep(c("MetaHD", "MetaHD_est.wscor", "Random", "Fixed"),
each = N),
levels=c("MetaHD", "MetaHD_est.wscor", "Random", "Fixed")),
Difference = c( true_values- model_MetaHD$estimate,
true_values- model_MetaHD_est.wscor$estimate,
true_values -model_random$estimate,
true_values -model_fixed$estimate))
# Create the boxplots
ggplot(df, aes(x = Model, y = Difference, fill = Model)) +
geom_boxplot() +
geom_hline(yintercept = 0, linetype = "dashed") +
scale_fill_manual(values = c("darkgreen", "red", "blue", "purple")) +
labs(title = "Differences between true and estimated values",
x = "Model",
y = "Difference (True value - Estiamted values)")