
ASPAR_KR–what is to come?
How do we move forward from the conference?
11/8/23
Last week
I presented the first part of my thesis at the ASPAR_KR conference.
There was limited discussion afterwards:
Unclearity about the motivation for FAIRDS.
Minimum information working group.
- Suggested by Paul, Mike and Johanna.
Interest during the conference
Martin Weichert.
Jianhua Zhang.
Now we need to move forward
Most important thing in my opinion
Agreeing on minimal information templates.
Minimum information
Working group
- Decision from experts.
- Members
- Bas.
- Paul.
- Mike.
- \(\dots\)?
Developing ASPAR_KR with Jasper
- Improve file upload.
- Use FAIRDS from
R. - More features?
Promoting the FAIRDS/ASPAR_KR
- Meetings
- Martin Weichert.
Researching the resistance of many lab-isolates of Aspergillus. - Christopher Wandabwa.
Researching metagenomes in fermented milk over geographical scales. - Do you know anyone else?
- Martin Weichert.
How will I accomplish this?
Timeline
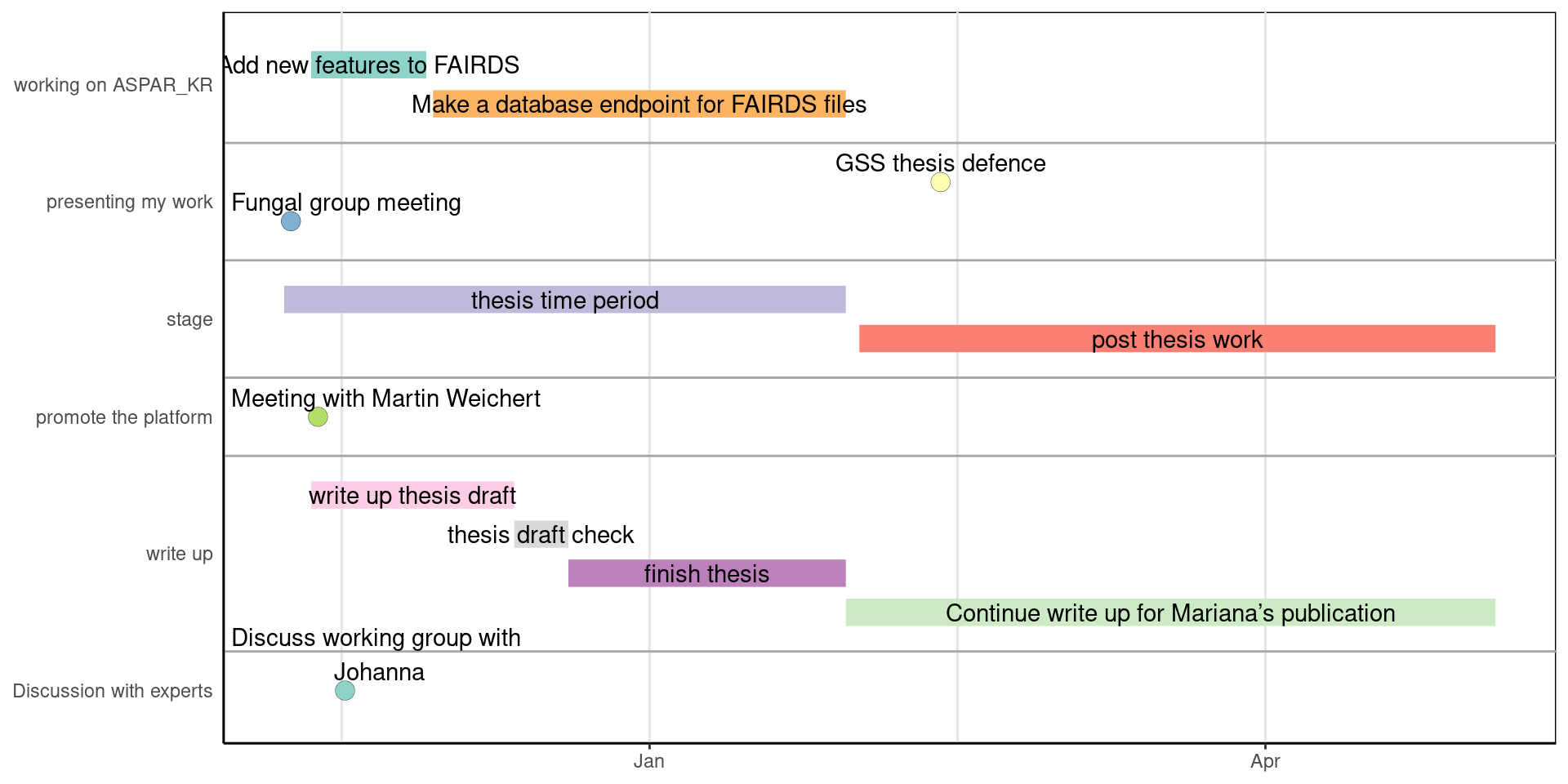
Planning of the remainer of the thesis.
A note on incentives and use cases
Jasper told me that his programme can only be promoted with a “big stick”.
To work with the UNLOCK programme, you must use fairds.
After using
FAIRDSpeople can make automated analyses.
How does this translate to our users?
Some authority could mandate the usage of FAIRDS/ASPAR_KR in collaborations.
An
Rpackage could be developed for most common analysis use cases and solve them automatically.
Questions?
Thanks for
Jasper Koehorst and Bart Nijsen.
Helping me contribute toFAIRDS.Mariana and Anna
Supervision during my stay at the genetics department.