3.7 Two Discrete
3.7.1 Distance Metrics
Some consider distance is not a correlation metric because it isn’t unit independent (i.e., if you scale the distance, the metrics will change), but it’s still a useful proxy. Distance metrics are more likely to be used for similarity measure.
Euclidean Distance
Manhattan Distance
Chessboard Distance
Minkowski Distance
Canberra Distance
Hamming Distance
Cosine Distance
Sum of Absolute Distance
Sum of Squared Distance
Mean-Absolute Error
3.7.2 Statistical Metrics
3.7.2.1 Chi-squared test
3.7.2.1.2 Cramer’s V
- between nominal categorical variables (no natural order)
\[ \text{Cramer's V} = \sqrt{\frac{\chi^2/n}{\min(c-1,r-1)}} \]
where
\(\chi^2\) = Chi-square statistic
\(n\) = sample size
\(r\) = # of rows
\(c\) = # of columns
library('lsr')
n = 100 # (sample size)
set.seed(1)
data = data.frame(A = sample(1:5, replace = TRUE, size = n),
B = sample(1:6, replace = TRUE, size = n))
cramersV(data$A, data$B)
#> [1] 0.1944616Alternatively,
ncchisqnoncentral Chi-squarenchisqadjAdjusted noncentral Chi-squarefisherFisher Z transformationfisheradjbias correction Fisher z transformation
3.7.3 Ordinal Association (Rank correlation)
- Good with non-linear relationship
3.7.3.1 Ordinal and Nominal
n = 100 # (sample size)
set.seed(1)
dt = table(data.frame(
A = sample(1:4, replace = TRUE, size = n), # ordinal
B = sample(1:3, replace = TRUE, size = n) # nominal
))
dt
#> B
#> A 1 2 3
#> 1 7 11 9
#> 2 11 6 14
#> 3 7 11 4
#> 4 6 4 103.7.3.2 Two Ordinal
n = 100 # (sample size)
set.seed(1)
dt = table(data.frame(
A = sample(1:4, replace = TRUE, size = n), # ordinal
B = sample(1:3, replace = TRUE, size = n) # ordinal
))
dt
#> B
#> A 1 2 3
#> 1 7 11 9
#> 2 11 6 14
#> 3 7 11 4
#> 4 6 4 103.7.3.2.4 Yule’s Q and Y
- 2 ordinal variables
Special version \((2 \times 2)\) of the Goodman Kruskal’s Gamma coefficient.
| Variable 1 | ||
|---|---|---|
| Variable 2 | a | b |
| c | d |
\[ \text{Yule's Q} = \frac{ad - bc}{ad + bc} \]
We typically use Yule’s \(Q\) in practice while Yule’s Y has the following relationship with \(Q\).
\[ \text{Yule's Y} = \frac{\sqrt{ad} - \sqrt{bc}}{\sqrt{ad} + \sqrt{bc}} \]
\[ Q = \frac{2Y}{1 + Y^2} \]
\[ Y = \frac{1 = \sqrt{1-Q^2}}{Q} \]
3.7.3.2.5 Tetrachoric Correlation
- is a special case of Polychoric Correlation when both variables are binary
library(psych)
n = 100 # (sample size)
data = data.frame(A = sample(c(0, 1), replace = TRUE, size = n),
B = sample(c(0, 1), replace = TRUE, size = n))
#view table
head(data)
#> A B
#> 1 1 0
#> 2 1 0
#> 3 0 0
#> 4 1 0
#> 5 1 0
#> 6 1 0
table(data)
#> B
#> A 0 1
#> 0 21 23
#> 1 34 22
#calculate tetrachoric correlation
tetrachoric(data)
#> Call: tetrachoric(x = data)
#> tetrachoric correlation
#> A B
#> A 1.0
#> B -0.2 1.0
#>
#> with tau of
#> A B
#> -0.15 0.133.7.3.2.6 Polychoric Correlation
- between ordinal categorical variables (natural order).
- Assumption: Ordinal variable is a discrete representation of a latent normally distributed continuous variable. (Income = low, normal, high).
library(polycor)
n = 100 # (sample size)
data = data.frame(A = sample(1:4, replace = TRUE, size = n),
B = sample(1:6, replace = TRUE, size = n))
head(data)
#> A B
#> 1 1 3
#> 2 1 1
#> 3 3 5
#> 4 2 3
#> 5 3 5
#> 6 4 4
#calculate polychoric correlation between ratings
polychor(data$A, data$B)
#> [1] 0.016079823.7.4 Summary
library(tidyverse)
data("mtcars")
df = mtcars %>%
dplyr::select(cyl, vs, carb)
df_factor = df %>%
dplyr::mutate(
cyl = factor(cyl),
vs = factor(vs),
carb = factor(carb)
)
# summary(df)
str(df)
#> 'data.frame': 32 obs. of 3 variables:
#> $ cyl : num 6 6 4 6 8 6 8 4 4 6 ...
#> $ vs : num 0 0 1 1 0 1 0 1 1 1 ...
#> $ carb: num 4 4 1 1 2 1 4 2 2 4 ...
str(df_factor)
#> 'data.frame': 32 obs. of 3 variables:
#> $ cyl : Factor w/ 3 levels "4","6","8": 2 2 1 2 3 2 3 1 1 2 ...
#> $ vs : Factor w/ 2 levels "0","1": 1 1 2 2 1 2 1 2 2 2 ...
#> $ carb: Factor w/ 6 levels "1","2","3","4",..: 4 4 1 1 2 1 4 2 2 4 ...Get the correlation table for continuous variables only
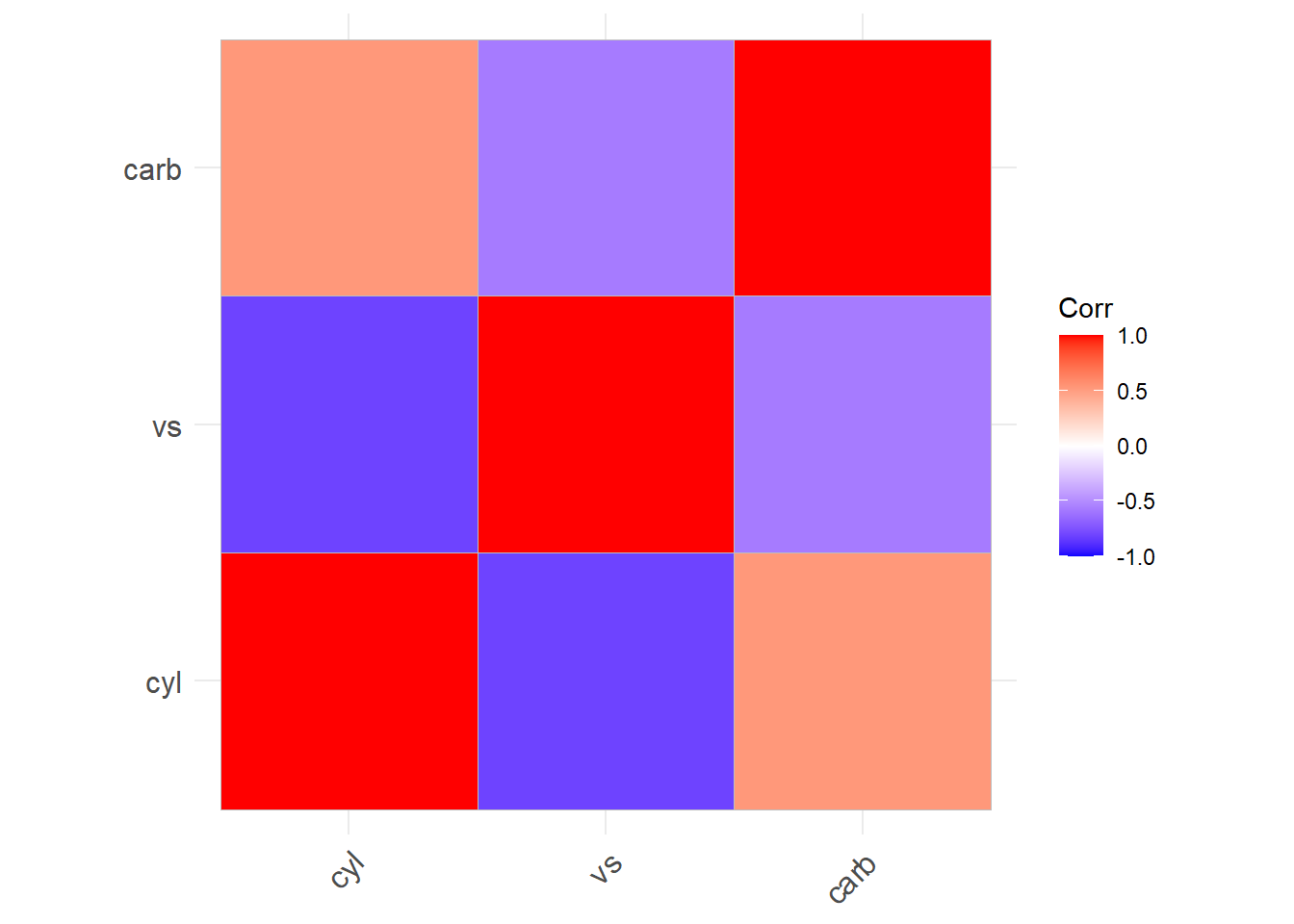
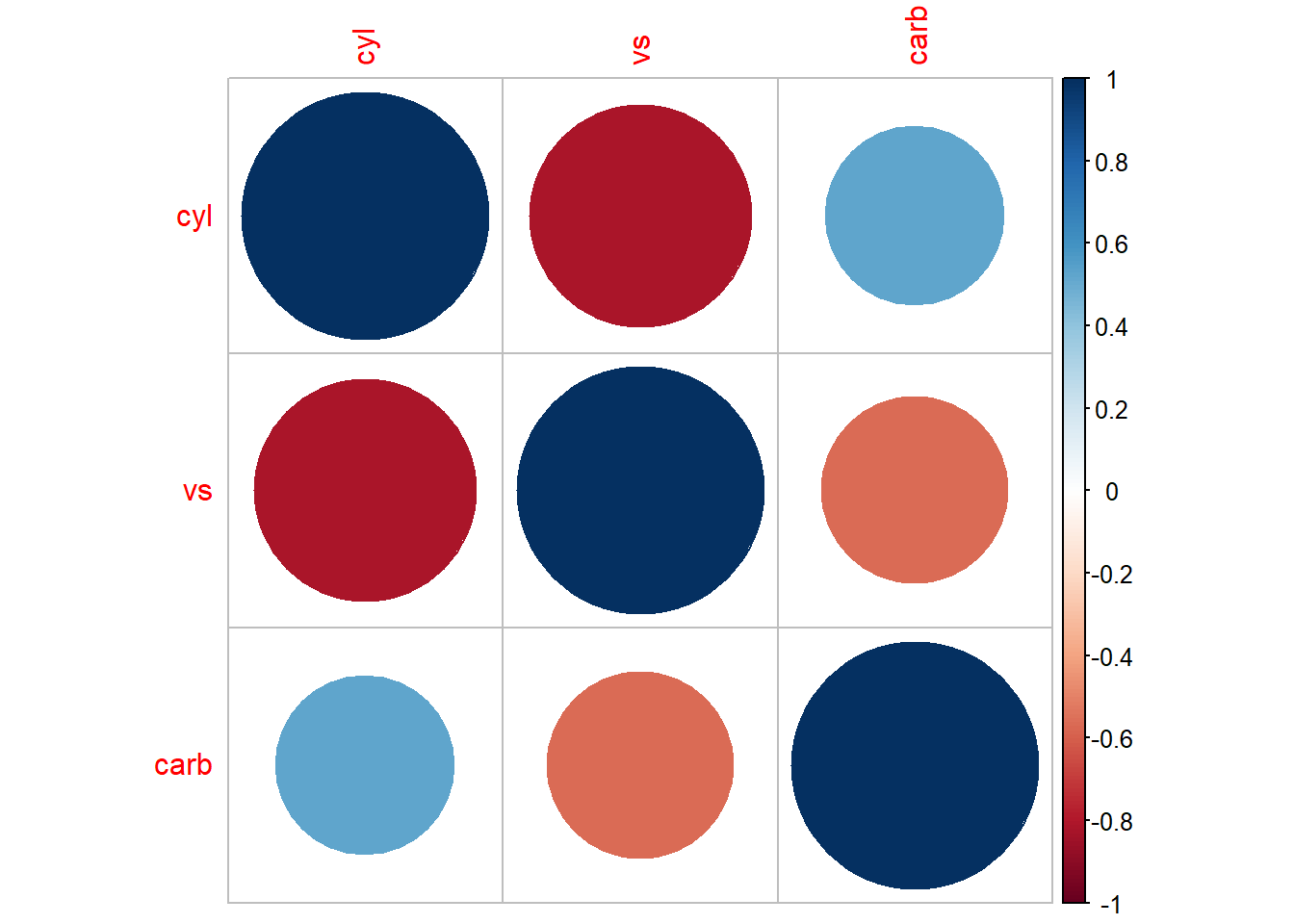
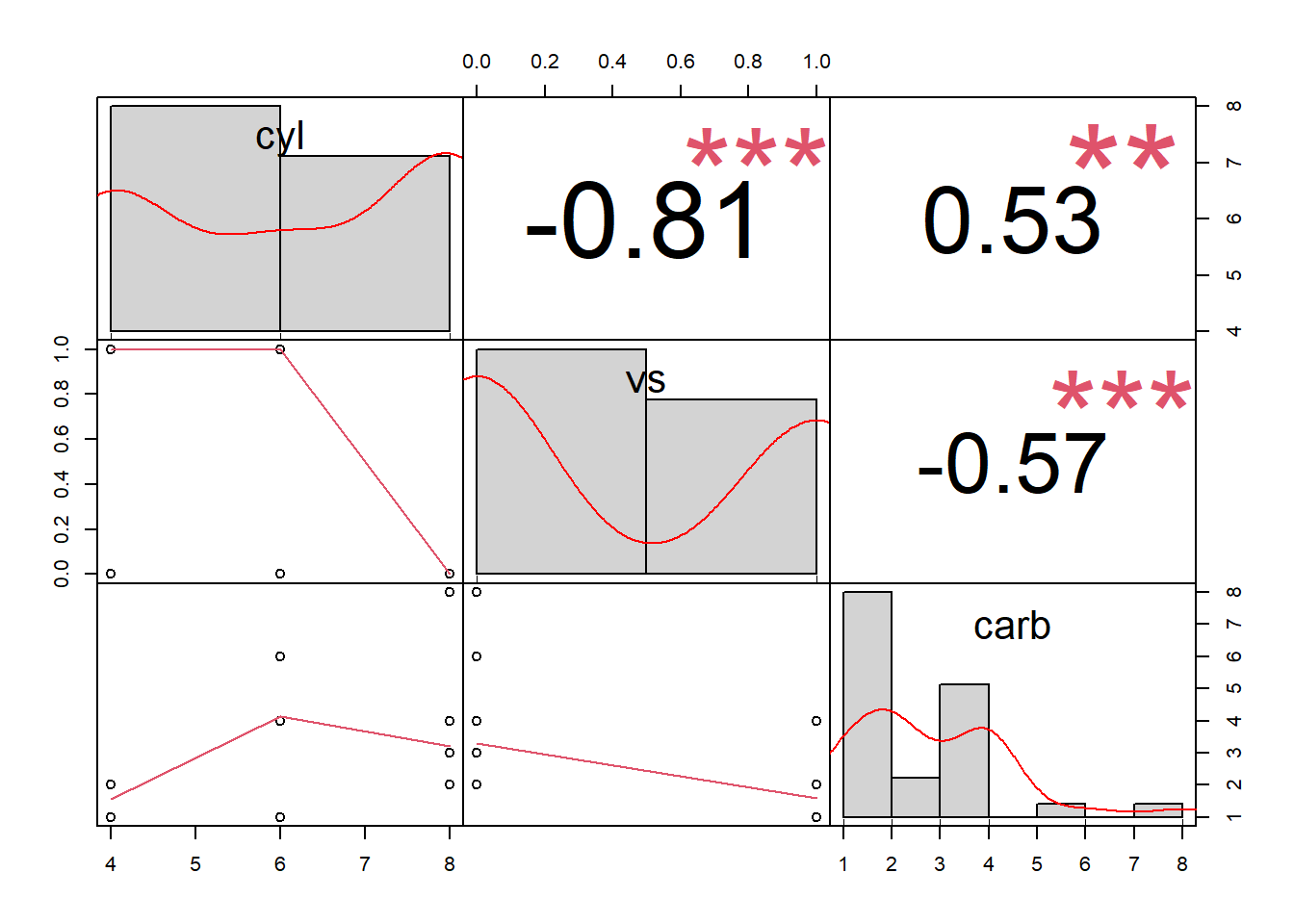
cor(df)
#> cyl vs carb
#> cyl 1.0000000 -0.8108118 0.5269883
#> vs -0.8108118 1.0000000 -0.5696071
#> carb 0.5269883 -0.5696071 1.0000000
# only complete obs
# cor(df, use = "complete.obs")Alternatively, you can also have the
Hmisc::rcorr(as.matrix(df), type = "pearson")
#> cyl vs carb
#> cyl 1.00 -0.81 0.53
#> vs -0.81 1.00 -0.57
#> carb 0.53 -0.57 1.00
#>
#> n= 32
#>
#>
#> P
#> cyl vs carb
#> cyl 0.0000 0.0019
#> vs 0.0000 0.0007
#> carb 0.0019 0.0007| cyl | vs | carb | |
|---|---|---|---|
| cyl | 1 | . | . |
| vs | −.81 | 1 | . |
| carb | .53 | −.57 | 1 |
Different comparison between different correlation between different types of variables (i.e., continuous vs. categorical) can be problematic. Moreover, the problem of detecting non-linear vs. linear relationship/correlation is another one. Hence, a solution is that using mutual information from information theory (i.e., knowing one variable can reduce uncertainty about the other).
To implement mutual information, we have the following approximations
\[ \downarrow \text{prediction error} \approx \downarrow \text{uncertainty} \approx \downarrow \text{association strength} \]
More specifically, following the X2Y metric, we have the following steps:
Predict \(y\) without \(x\) (i.e., baseline model)
Average of \(y\) when \(y\) is continuous
Most frequent value when \(y\) is categorical
Predict \(y\) with \(x\) (e.g., linear, random forest, etc.)
Calculate the prediction error difference between 1 and 2
To have a comprehensive table that could handle
continuous vs. continuous
categorical vs. continuous
continuous vs. categorical
categorical vs. categorical
the suggested model would be Classification and Regression Trees (CART). But we can certainly use other models as well.
The downfall of this method is that you might suffer
- Symmetry: \((x,y) \neq (y,x)\)
- Comparability : Different pair of comparison might use different metrics (e.g., misclassification error vs. MAE)
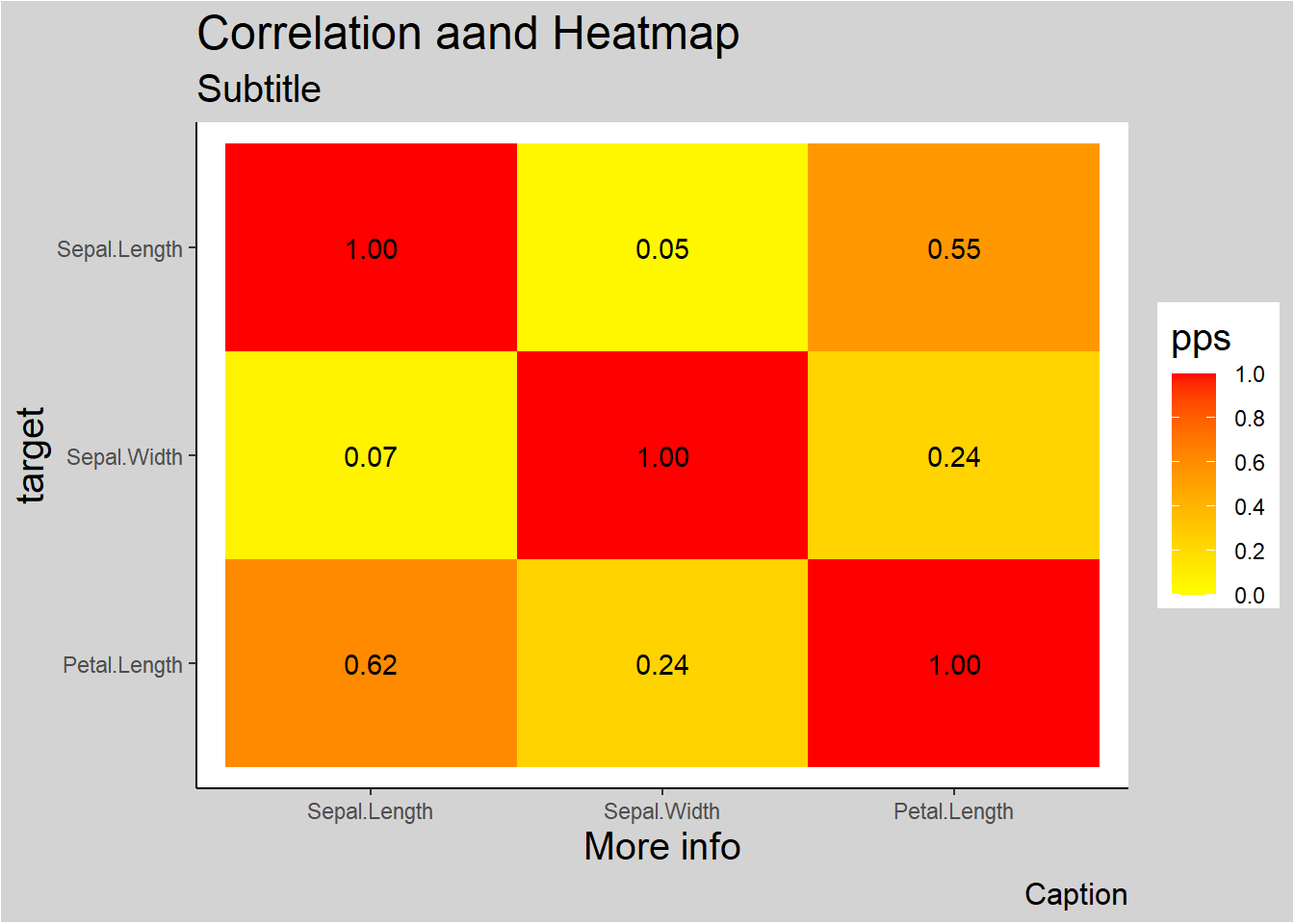
library(ppsr)
iris <- iris %>%
select(1:3)
# ppsr::score_df(iris) # if you want a dataframe
ppsr::score_matrix(iris,
do_parallel = TRUE,
n_cores = parallel::detectCores() / 2)
#> Sepal.Length Sepal.Width Petal.Length
#> Sepal.Length 1.00000000 0.04632352 0.5491398
#> Sepal.Width 0.06790301 1.00000000 0.2376991
#> Petal.Length 0.61608360 0.24263851 1.0000000
# if you want a similar correlation matrix
ppsr::score_matrix(df,
do_parallel = TRUE,
n_cores = parallel::detectCores() / 2)
#> cyl vs carb
#> cyl 1.00000000 0.3982789 0.2092533
#> vs 0.02514286 1.0000000 0.2000000
#> carb 0.30798148 0.2537309 1.00000003.7.5 Visualization
Alternatively,
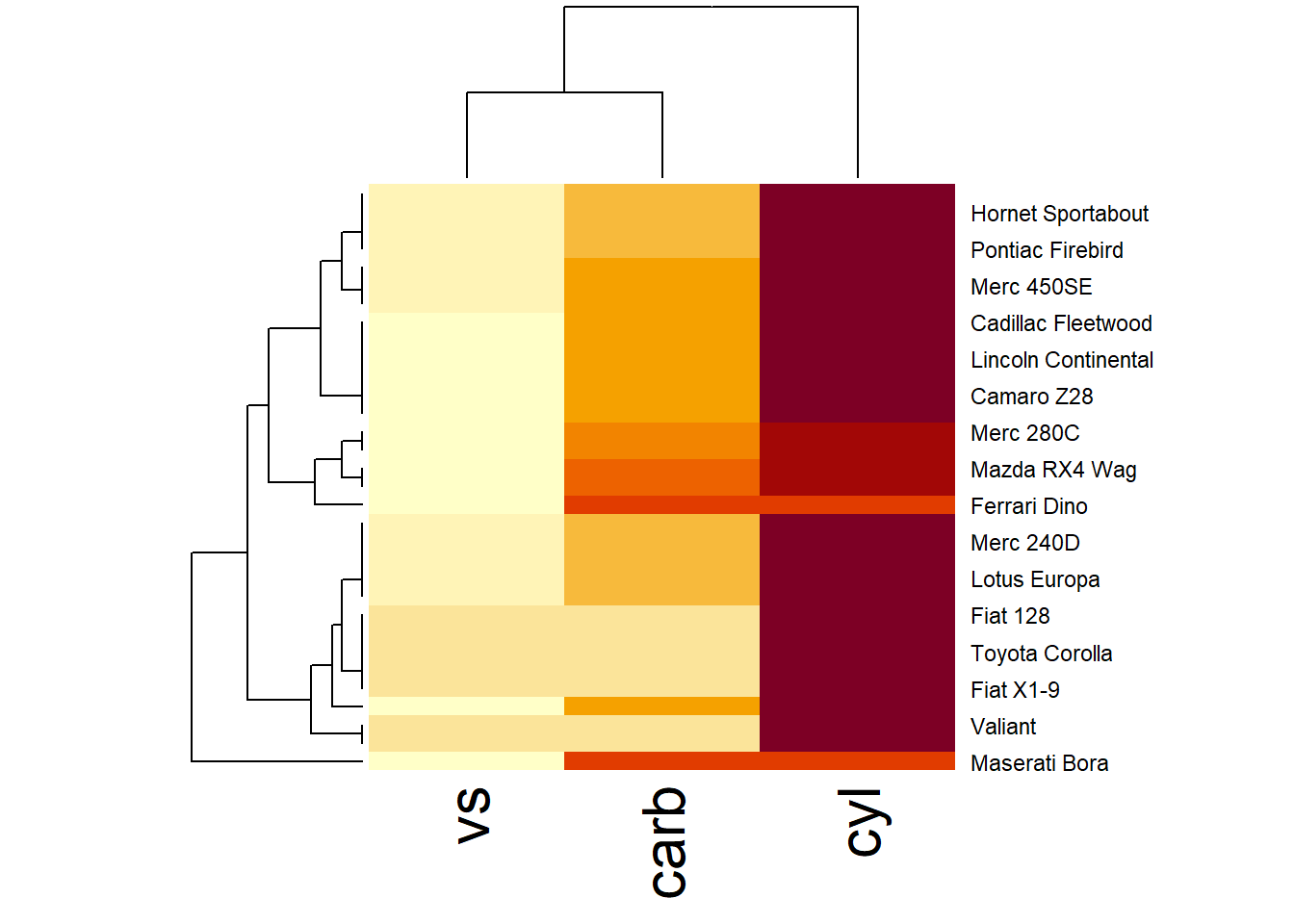
More general form,
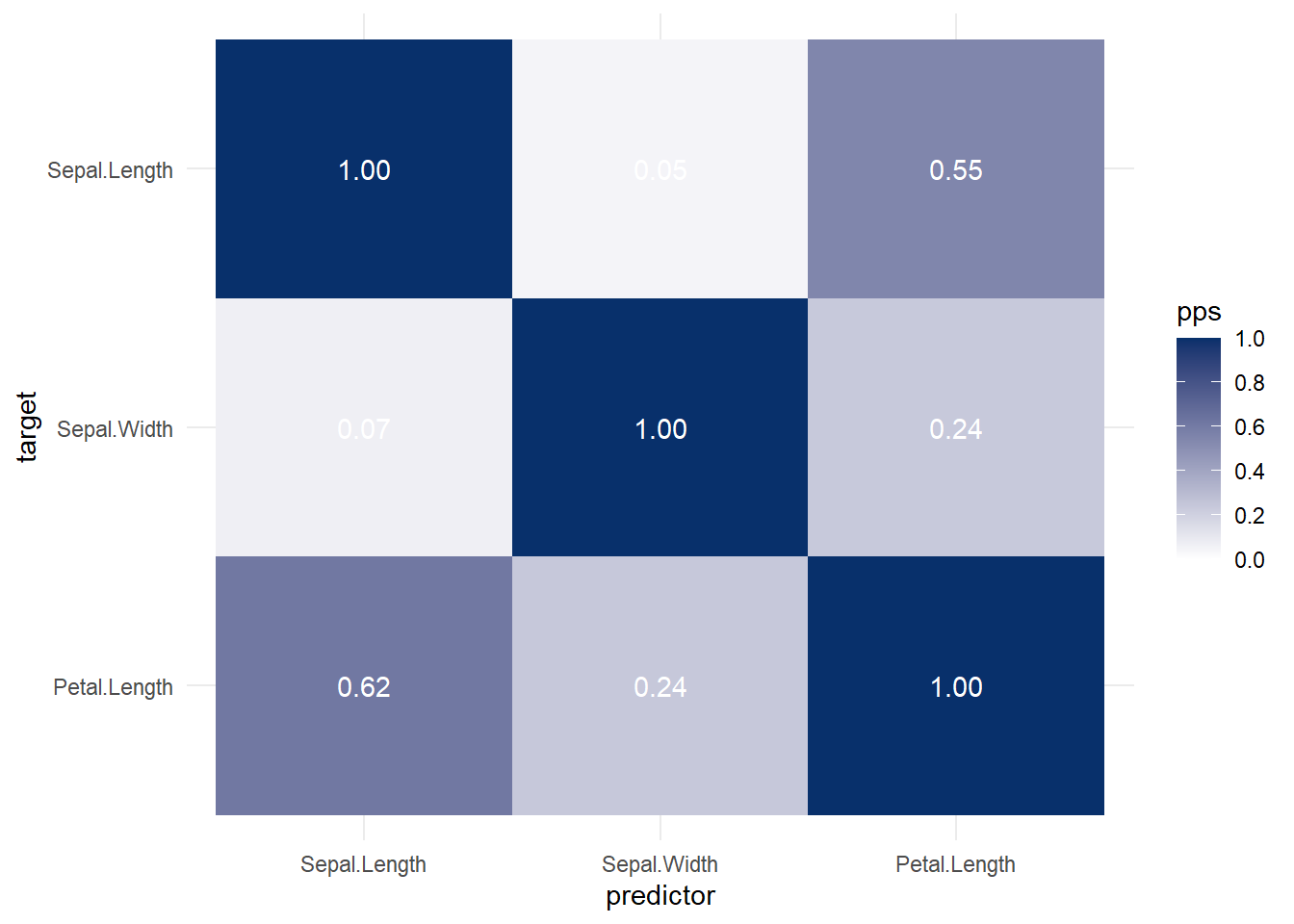
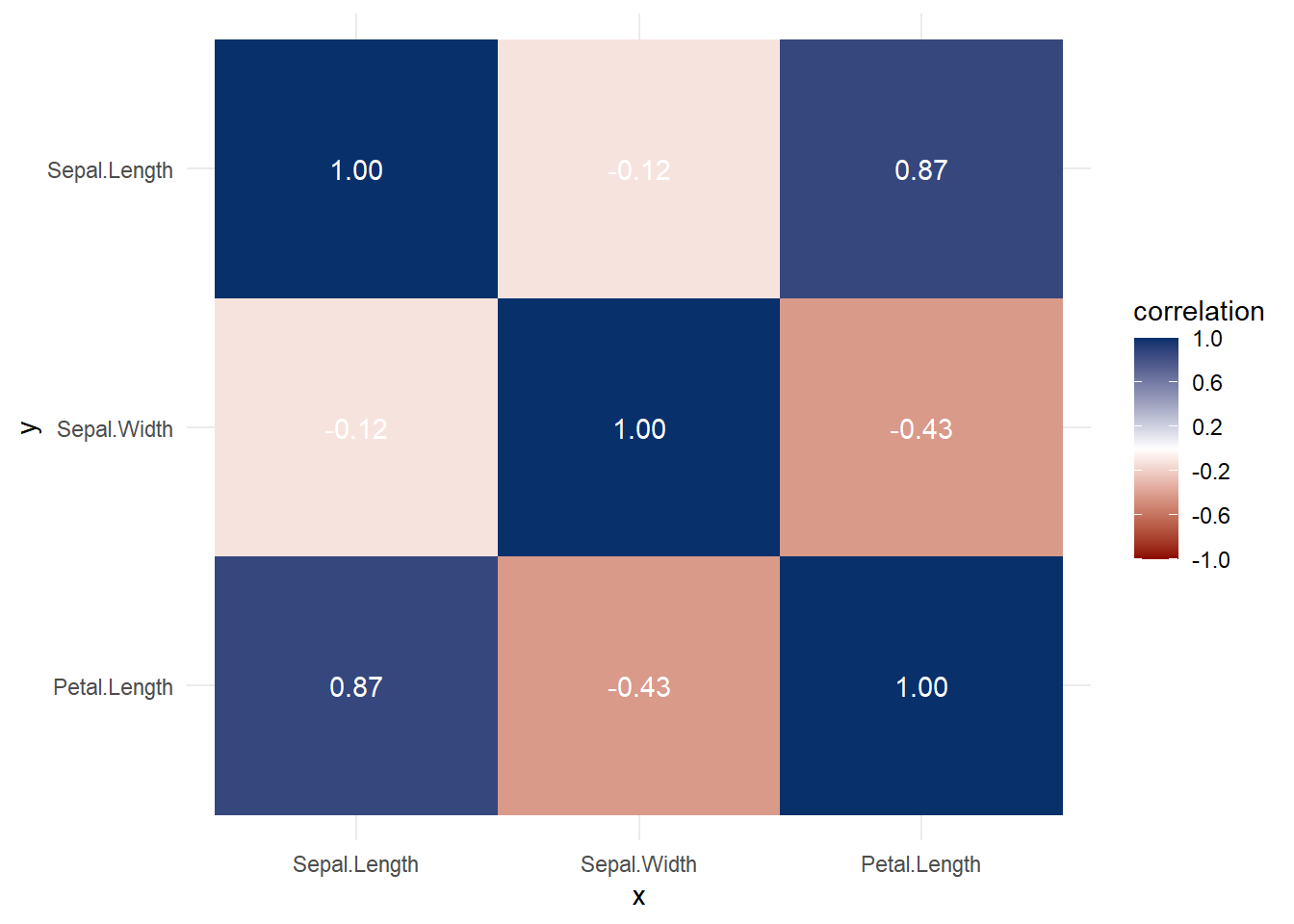
Both heat map and correlation at the same time
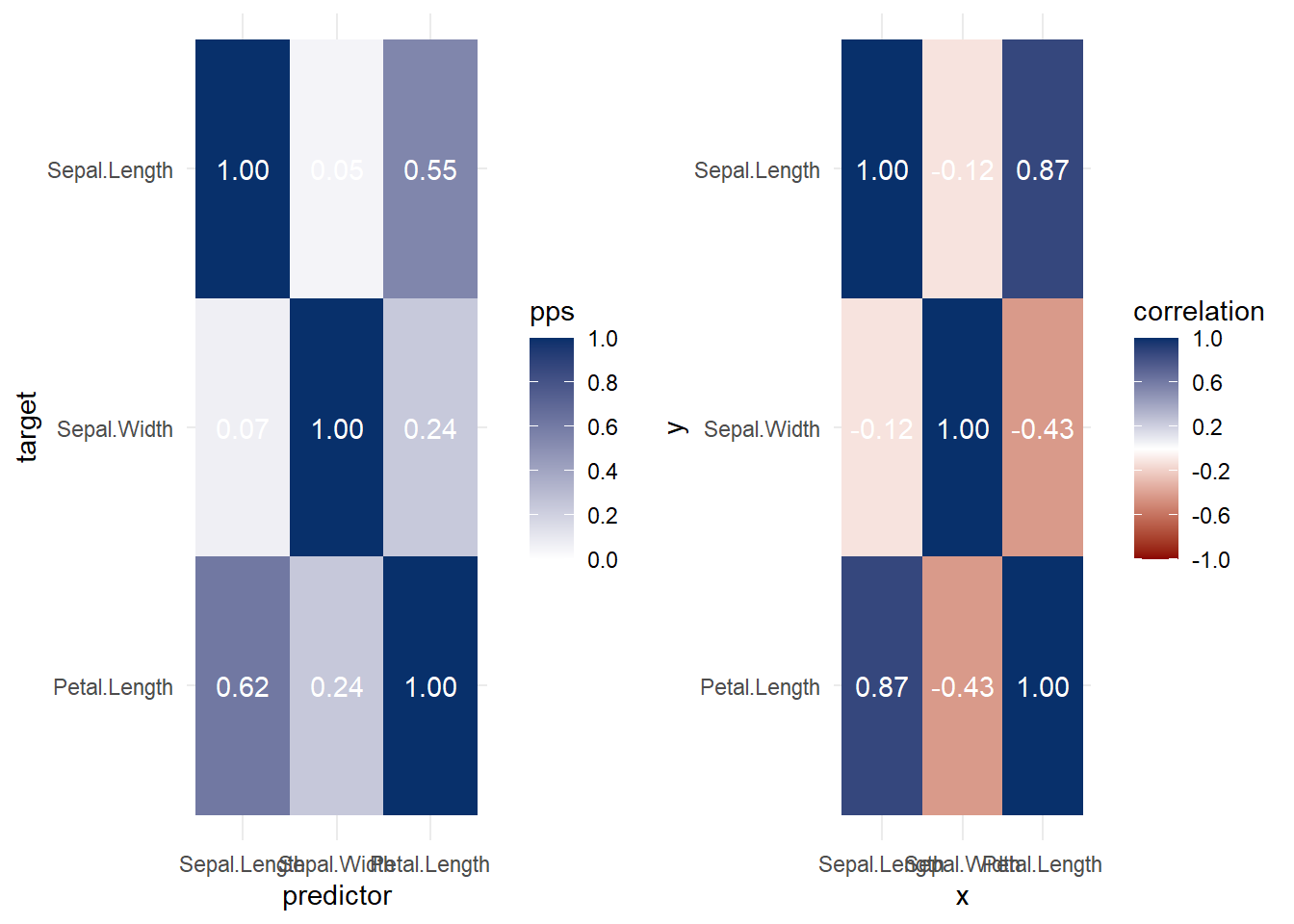
More elaboration with ggplot2
ppsr::visualize_pps(
df = iris,
color_value_high = 'red',
color_value_low = 'yellow',
color_text = 'black'
) +
ggplot2::theme_classic() +
ggplot2::theme(plot.background =
ggplot2::element_rect(fill = "lightgrey")) +
ggplot2::theme(title = ggplot2::element_text(size = 15)) +
ggplot2::labs(
title = 'Correlation aand Heatmap',
subtitle = 'Subtitle',
caption = 'Caption',
x = 'More info'
)